Simplifications and approximations in a single-gene circuit modeling
- PMID: 38822072
- PMCID: PMC11143231
- DOI: 10.1038/s41598-024-63265-8
Simplifications and approximations in a single-gene circuit modeling
Abstract
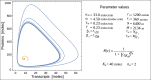
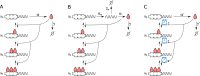
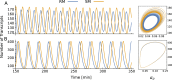
The absence of detailed knowledge about regulatory interactions makes the use of phenomenological assumptions mandatory in cell biology modeling. Furthermore, the challenges associated with the analysis of these models compel the implementation of mathematical approximations. However, the constraints these methods introduce to biological interpretation are sometimes neglected. Consequently, understanding these restrictions is a very important task for systems biology modeling. In this article, we examine the impact of such simplifications, taking the case of a single-gene autoinhibitory circuit; however, our conclusions are not limited solely to this instance. We demonstrate that models grounded in the same biological assumptions but described at varying levels of detail can lead to different outcomes, that is, different and contradictory phenotypes or behaviors. Indeed, incorporating specific molecular processes like translation and elongation into the model can introduce instabilities and oscillations not seen when these processes are assumed to be instantaneous. Furthermore, incorporating a detailed description of promoter dynamics, usually described by a phenomenological regulatory function, can lead to instability, depending on the cooperative binding mechanism that is acting. Consequently, although the use of a regulating function facilitates model analysis, it may mask relevant aspects of the system's behavior. In particular, we observe that the two cooperative binding mechanisms, both compatible with the same sigmoidal function, can lead to different phenotypes, such as transcriptional oscillations with different oscillation frequencies.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
The art of modeling gene regulatory circuits.NPJ Syst Biol Appl. 2024 May 29;10(1):60. doi: 10.1038/s41540-024-00380-2. NPJ Syst Biol Appl. 2024. PMID: 38811585 Free PMC article.
-
A Model for the Spatiotemporal Design of Gene Regulatory Circuits †.ACS Synth Biol. 2019 Sep 20;8(9):2007-2016. doi: 10.1021/acssynbio.9b00022. Epub 2019 Aug 30. ACS Synth Biol. 2019. PMID: 31429541
-
Foundations for modeling the dynamics of gene regulatory networks: a multilevel-perspective review.J Bioinform Comput Biol. 2014 Feb;12(1):1330003. doi: 10.1142/S0219720013300037. Epub 2013 Dec 20. J Bioinform Comput Biol. 2014. PMID: 24467752 Review.
-
Gradient descent optimization in gene regulatory pathways.PLoS One. 2010 Sep 3;5(9):e12475. doi: 10.1371/journal.pone.0012475. PLoS One. 2010. PMID: 20838430 Free PMC article.
-
Mathematical formalisms based on approximated kinetic representations for modeling genetic and metabolic pathways.Biotechnol Genet Eng Rev. 2008;25:1-40. doi: 10.5661/bger-25-1. Biotechnol Genet Eng Rev. 2008. PMID: 21412348 Review.
References
MeSH terms
LinkOut - more resources
Full Text Sources

