ScAnalyzer: an image processing tool to monitor plant disease symptoms and pathogen spread in Arabidopsis thaliana leaves
- PMID: 38822355
- PMCID: PMC11141064
- DOI: 10.1186/s13007-024-01213-3
ScAnalyzer: an image processing tool to monitor plant disease symptoms and pathogen spread in Arabidopsis thaliana leaves
Abstract
Background: Plants are known to be infected by a wide range of pathogenic microbes. To study plant diseases caused by microbes, it is imperative to be able to monitor disease symptoms and microbial colonization in a quantitative and objective manner. In contrast to more traditional measures that use manual assignments of disease categories, image processing provides a more accurate and objective quantification of plant disease symptoms. Besides monitoring disease symptoms, computational image processing provides additional information on the spatial localization of pathogenic microbes in different plant tissues.
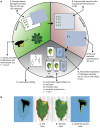
Results: Here we report on an image analysis tool called ScAnalyzer to monitor disease symptoms and bacterial spread in Arabidopsis thaliana leaves. Thereto, detached leaves are assembled in a grid and scanned, which enables automated separation of individual samples. A pixel color threshold is used to segment healthy (green) from chlorotic (yellow) leaf areas. The spread of luminescence-tagged bacteria is monitored via light-sensitive films, which are processed in a similar manner as the leaf scans. We show that this tool is able to capture previously identified differences in susceptibility of the model plant A. thaliana to the bacterial pathogen Xanthomonas campestris pv. campestris. Moreover, we show that the ScAnalyzer pipeline provides a more detailed assessment of bacterial spread within plant leaves than previously used methods. Finally, by combining the disease symptom values with bacterial spread values from the same leaves, we show that bacterial spread precedes visual disease symptoms.
Conclusion: Taken together, we present an automated script to monitor plant disease symptoms and microbial spread in A. thaliana leaves. The freely available software ( https://github.com/MolPlantPathology/ScAnalyzer ) has the potential to standardize the analysis of disease assays between different groups.
Keywords: Arabidopsis thaliana; Xanthomonas campestris; Bioluminescence; Black rot disease; Digital phenotyping; Image processing; Plant disease.
© 2024. The Author(s).
Conflict of interest statement
HvdB is employed by Keygene N.V., Wageningen, The Netherlands. Keygene was not involved in any part of this study.
Figures



References
-
- Bock C, Poole G, Parker P, Gottwald T. Plant disease severity estimated visually, by digital photography and image analysis, and by hyperspectral imaging. Crit Rev Plant Sci. 2010;29(2):59–107. doi: 10.1080/07352681003617285. - DOI
-
- Lindow S. Estimating disease severity of single plants. Phytopathology. 1983;73(11):1576–81. doi: 10.1094/Phyto-73-1576. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

