METTL1-mediated tRNA m7G methylation and translational dysfunction restricts breast cancer tumorigenesis by fueling cell cycle blockade
- PMID: 38822363
- PMCID: PMC11140866
- DOI: 10.1186/s13046-024-03076-x
METTL1-mediated tRNA m7G methylation and translational dysfunction restricts breast cancer tumorigenesis by fueling cell cycle blockade
Abstract
Background: RNA modifications of transfer RNAs (tRNAs) are critical for tRNA function. Growing evidence has revealed that tRNA modifications are related to various disease processes, including malignant tumors. However, the biological functions of methyltransferase-like 1 (METTL1)-regulated m7G tRNA modifications in breast cancer (BC) remain largely obscure.
Methods: The biological role of METTL1 in BC progression were examined by cellular loss- and gain-of-function tests and xenograft models both in vitro and in vivo. To investigate the change of m7G tRNA modification and mRNA translation efficiency in BC, m7G-methylated tRNA immunoprecipitation sequencing (m7G tRNA MeRIP-seq), Ribosome profiling sequencing (Ribo-seq), and polysome-associated mRNA sequencing were performed. Rescue assays were conducted to decipher the underlying molecular mechanisms.
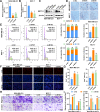
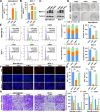
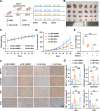
Results: The tRNA m7G methyltransferase complex components METTL1 and WD repeat domain 4 (WDR4) were down-regulated in BC tissues at both the mRNA and protein levels. Functionally, METTL1 inhibited BC cell proliferation, and cell cycle progression, relying on its enzymatic activity. Mechanistically, METTL1 increased m7G levels of 19 tRNAs to modulate the translation of growth arrest and DNA damage 45 alpha (GADD45A) and retinoblastoma protein 1 (RB1) in a codon-dependent manner associated with m7G. Furthermore, in vivo experiments showed that overexpression of METTL1 enhanced the anti-tumor effectiveness of abemaciclib, a cyclin-dependent kinases 4 and 6 (CDK4/6) inhibitor.
Conclusion: Our study uncovered the crucial tumor-suppressive role of METTL1-mediated tRNA m7G modification in BC by promoting the translation of GADD45A and RB1 mRNAs, selectively blocking the G2/M phase of the cell cycle. These findings also provided a promising strategy for improving the therapeutic benefits of CDK4/6 inhibitors in the treatment of BC patients.
Keywords: Breast cancer; CDK4/6; Cell cycle; GADD45A; METTL1.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

