Development and validation of a prognostic model for assessing long COVID risk following Omicron wave-a large population-based cohort study
- PMID: 38822405
- PMCID: PMC11140920
- DOI: 10.1186/s12985-024-02400-3
Development and validation of a prognostic model for assessing long COVID risk following Omicron wave-a large population-based cohort study
Abstract
Background: Long coronavirus disease (COVID) after COVID-19 infection is continuously threatening the health of people all over the world. Early prediction of the risk of Long COVID in hospitalized patients will help clinical management of COVID-19, but there is still no reliable and effective prediction model.
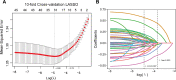
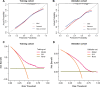
Methods: A total of 1905 hospitalized patients with COVID-19 infection were included in this study, and their Long COVID status was followed up 4-8 weeks after discharge. Univariable and multivariable logistic regression analysis were used to determine the risk factors for Long COVID. Patients were randomly divided into a training cohort (70%) and a validation cohort (30%), and factors for constructing the model were screened using Lasso regression in the training cohort. Visualize the Long COVID risk prediction model using nomogram. Evaluate the performance of the model in the training and validation cohort using the area under the curve (AUC), calibration curve, and decision curve analysis (DCA).
Results: A total of 657 patients (34.5%) reported that they had symptoms of long COVID. The most common symptoms were fatigue or muscle weakness (16.8%), followed by sleep difficulties (11.1%) and cough (9.5%). The risk prediction nomogram of age, diabetes, chronic kidney disease, vaccination status, procalcitonin, leukocytes, lymphocytes, interleukin-6 and D-dimer were included for early identification of high-risk patients with Long COVID. AUCs of the model in the training cohort and validation cohort are 0.762 and 0.713, respectively, demonstrating relatively high discrimination of the model. The calibration curve further substantiated the proximity of the nomogram's predicted outcomes to the ideal curve, the consistency between the predicted outcomes and the actual outcomes, and the potential benefits for all patients as indicated by DCA. This observation was further validated in the validation cohort.
Conclusions: We established a nomogram model to predict the long COVID risk of hospitalized patients with COVID-19, and proved its relatively good predictive performance. This model is helpful for the clinical management of long COVID.
Keywords: Long COVID; Nomogram; Omicron; Prediction.
© 2024. The Author(s).
Conflict of interest statement
All authors have no conflict of interest.
Figures

Similar articles
-
Construction and validation of a machine learning-based nomogram: A tool to predict the risk of getting severe coronavirus disease 2019 (COVID-19).Immun Inflamm Dis. 2021 Jun;9(2):595-607. doi: 10.1002/iid3.421. Epub 2021 Mar 13. Immun Inflamm Dis. 2021. PMID: 33713584 Free PMC article.
-
A Prediction Model for Prolonged Hospital Length of Stay (ProLOS) in Coronavirus Disease 2019 (COVID-19) Patients.Clin Lab. 2024 May 1;70(5). doi: 10.7754/Clin.Lab.2023.231203. Clin Lab. 2024. PMID: 38747926
-
Development and validation of a nomogram to assess the occurrence of liver dysfunction in patients with COVID-19 pneumonia in the ICU.BMC Infect Dis. 2025 Mar 10;25(1):332. doi: 10.1186/s12879-025-10684-1. BMC Infect Dis. 2025. PMID: 40065225 Free PMC article.
-
A novel combined nomogram for predicting severe acute lower respiratory tract infection in children hospitalized for RSV infection during the post-COVID-19 period.Front Immunol. 2024 Jul 24;15:1437834. doi: 10.3389/fimmu.2024.1437834. eCollection 2024. Front Immunol. 2024. PMID: 39114651 Free PMC article.
-
Development and Validation of a Nomogram for Assessing Survival in Patients With COVID-19 Pneumonia.Clin Infect Dis. 2021 Feb 16;72(4):652-660. doi: 10.1093/cid/ciaa963. Clin Infect Dis. 2021. PMID: 32649738 Free PMC article.
Cited by
-
Factors affecting the impact of COVID-19 vaccination on post COVID-19 conditions among adults: A systematic literature review.Hum Vaccin Immunother. 2025 Dec;21(1):2474772. doi: 10.1080/21645515.2025.2474772. Epub 2025 Mar 13. Hum Vaccin Immunother. 2025. PMID: 40079963 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

