Sarbecovirus disease susceptibility is conserved across viral and host models
- PMID: 38823688
- PMCID: PMC11225686
- DOI: 10.1016/j.virusres.2024.199399
Sarbecovirus disease susceptibility is conserved across viral and host models
Abstract
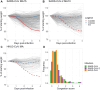
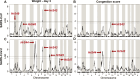
Coronaviruses have caused three severe epidemics since the start of the 21st century: SARS, MERS and COVID-19. The severity of the ongoing COVID-19 pandemic and increasing likelihood of future coronavirus outbreaks motivates greater understanding of factors leading to severe coronavirus disease. We screened ten strains from the Collaborative Cross mouse genetic reference panel and identified strains CC006/TauUnc (CC006) and CC044/Unc (CC044) as coronavirus-susceptible and resistant, respectively, as indicated by variable weight loss and lung congestion scores four days post-infection. We generated a genetic mapping population of 755 CC006xCC044 F2 mice and exposed the mice to one of three genetically distinct mouse-adapted coronaviruses: clade 1a SARS-CoV MA15 (n=391), clade 1b SARS-CoV-2 MA10 (n=274), and clade 2 HKU3-CoV MA (n=90). Quantitative trait loci (QTL) mapping in SARS-CoV MA15- and SARS-CoV-2 MA10-infected F2 mice identified genetic loci associated with disease severity. Specifically, we identified seven loci associated with variation in outcome following infection with either virus, including one, HrS43, that is present in both groups. Three of these QTL, including HrS43, were also associated with HKU3-CoV MA outcome. HrS43 overlaps with a QTL previously reported by our lab that is associated with SARS-CoV MA15 outcome in CC011xCC074 F2 mice and is also syntenic with a human chromosomal region associated with severe COVID-19 outcomes in humans GWAS. The results reported here provide: (a) additional support for the involvement of this locus in SARS-CoV MA15 infection, (b) the first conclusive evidence that this locus is associated with susceptibility across the Sarbecovirus subgenus, and (c) demonstration of the relevance of mouse models in the study of coronavirus disease susceptibility in humans.
Keywords: COVID-19; Collaborative Cross; Coronavirus; Genetics; Mouse models; QTL; SARS; SARS-CoV; SARS-CoV-2.
Copyright © 2024 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: RSB and SRL are listed as inventors on a patent pertaining to the mouse-adapted SARS-CoV-2 (MA10) virus used in this study (Patent number: 11,225,508)
Figures



Similar articles
-
Genetic loci regulate Sarbecovirus pathogenesis: A comparison across mice and humans.Virus Res. 2024 Jun;344:199357. doi: 10.1016/j.virusres.2024.199357. Epub 2024 Mar 23. Virus Res. 2024. PMID: 38508400 Free PMC article.
-
Antibody tests for identification of current and past infection with SARS-CoV-2.Cochrane Database Syst Rev. 2022 Nov 17;11(11):CD013652. doi: 10.1002/14651858.CD013652.pub2. Cochrane Database Syst Rev. 2022. PMID: 36394900 Free PMC article.
-
CCR2 Signaling Restricts SARS-CoV-2 Infection.mBio. 2021 Dec 21;12(6):e0274921. doi: 10.1128/mBio.02749-21. Epub 2021 Nov 9. mBio. 2021. PMID: 34749524 Free PMC article.
-
Measures implemented in the school setting to contain the COVID-19 pandemic.Cochrane Database Syst Rev. 2022 Jan 17;1(1):CD015029. doi: 10.1002/14651858.CD015029. Cochrane Database Syst Rev. 2022. Update in: Cochrane Database Syst Rev. 2024 May 2;5:CD015029. doi: 10.1002/14651858.CD015029.pub2. PMID: 35037252 Free PMC article. Updated.
-
SARS-CoV-2-neutralising monoclonal antibodies for treatment of COVID-19.Cochrane Database Syst Rev. 2021 Sep 2;9(9):CD013825. doi: 10.1002/14651858.CD013825.pub2. Cochrane Database Syst Rev. 2021. PMID: 34473343 Free PMC article.
Cited by
-
Mouse Adapted Omicron BA.5 Induces A Fibrotic Lung Disease Phenotype in BALB/c Mice.bioRxiv [Preprint]. 2025 Jul 16:2025.07.16.665104. doi: 10.1101/2025.07.16.665104. bioRxiv. 2025. PMID: 40791410 Free PMC article. Preprint.
-
Animal models of Long Covid: A hit-and-run disease.Sci Transl Med. 2024 Nov 13;16(773):eado2104. doi: 10.1126/scitranslmed.ado2104. Epub 2024 Nov 13. Sci Transl Med. 2024. PMID: 39536118 Free PMC article. Review.
References
-
- ADAMS L.E., LEIST S.R., DINNON K.H., 3RD, WEST A., GULLY K.L., ANDERSON E.J., LOOME J.F., MADDEN E.A., POWERS J.M., SCHAFER A., SARKAR S., CASTILLO I.N., MARON J.S., MCNAMARA R.P., BERTERA H.L., ZWEIGERT M.R., HIGGINS J.S., HAMPTON B.K., PREMKUMAR L., ALTER G., MONTGOMERY S.A., BAXTER V.K., HEISE M.T., BARIC R.S. Fc-mediated pan-sarbecovirus protection after alphavirus vector vaccination. Cell Rep. 2023;42 - PMC - PubMed
-
- BROMAN K.W., WU H., SEN S., CHURCHILL G.A. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003;19:889–890. - PubMed
-
- CARLSON C.J., ALBERY G.F., MEROW C., TRISOS C.H., ZIPFEL C.M., ESKEW E.A., OLIVAL K.J., ROSS N., BANSAL S. Climate change increases cross-species viral transmission risk. Nature. 2022;607:555–562. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

