Structural basis for human mitochondrial tRNA maturation
- PMID: 38824131
- PMCID: PMC11144196
- DOI: 10.1038/s41467-024-49132-0
Structural basis for human mitochondrial tRNA maturation
Abstract
The human mitochondrial genome is transcribed into two RNAs, containing mRNAs, rRNAs and tRNAs, all dedicated to produce essential proteins of the respiratory chain. The precise excision of tRNAs by the mitochondrial endoribonucleases (mt-RNase), P and Z, releases all RNA species from the two RNA transcripts. The tRNAs then undergo 3'-CCA addition. In metazoan mitochondria, RNase P is a multi-enzyme assembly that comprises the endoribonuclease PRORP and a tRNA methyltransferase subcomplex. The requirement for this tRNA methyltransferase subcomplex for mt-RNase P cleavage activity, as well as the mechanisms of pre-tRNA 3'-cleavage and 3'-CCA addition, are still poorly understood. Here, we report cryo-EM structures that visualise four steps of mitochondrial tRNA maturation: 5' and 3' tRNA-end processing, methylation and 3'-CCA addition, and explain the defined sequential order of the tRNA processing steps. The methyltransferase subcomplex recognises the pre-tRNA in a distinct mode that can support tRNA-end processing and 3'-CCA addition, likely resulting from an evolutionary adaptation of mitochondrial tRNA maturation complexes to the structurally-fragile mitochondrial tRNAs. This subcomplex can also ensure a tRNA-folding quality-control checkpoint before the sequential docking of the maturation enzymes. Altogether, our study provides detailed molecular insight into RNA-transcript processing and tRNA maturation in human mitochondria.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

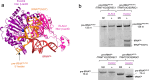




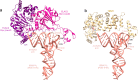
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

