Phenotypic and molecular characterization of extended spectrum- and metallo- beta lactamase producing Pseudomonas aeruginosa clinical isolates from Egypt
- PMID: 38824475
- PMCID: PMC11621155
- DOI: 10.1007/s15010-024-02297-8
Phenotypic and molecular characterization of extended spectrum- and metallo- beta lactamase producing Pseudomonas aeruginosa clinical isolates from Egypt
Abstract
Background: Antimicrobial resistance among Pseudomonas aeruginosa (P. aeruginosa), a leading cause of nosocomial infections worldwide, is escalating. This study investigated the prevalence of extended-spectrum β-lactamases (ESBLs) and metallo-β-lactamases (MBLs) among 104 P. aeruginosa clinical isolates from Alexandria Main University Hospital, Alexandria, Egypt.
Methods: Antimicrobial susceptibility testing was performed using agar dilution technique, or broth microdilution method in case of colistin. ESBL and MBL prevalence was assessed phenotypically and genotypically using polymerase chain reaction (PCR). The role of plasmids in mediating resistance to extended-spectrum β-lactams was studied via transformation technique using plasmids isolated from ceftazidime-resistant isolates.
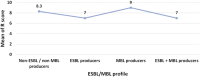
Results: Antimicrobial susceptibility testing revealed alarming resistance rates to carbapenems, cephalosporins, and fluoroquinolones. Using PCR as the gold standard, phenotypic methods underestimated ESBL production while overestimating MBL production. Eighty-five isolates (81.7%) possessed only ESBL encoding genes, among which 69 isolates harbored a single ESBL gene [blaOXA-10 (n = 67) and blaPER (n = 2)]. Four ESBL-genotype combinations were detected: blaPER + blaOXA-10 (n = 8), blaVEB-1 + blaOXA-10 (n = 6), blaPSE + blaOXA-10 (n = 1), and blaPER + blaVEB-1 + blaOXA-10 (n = 1). Three isolates (2.9%) possessed only the MBL encoding gene blaVIM. Three ESBL + MBL- genotype combinations: blaOXA-10 + blaAIM, blaOXA-10 + blaVIM, and blaPER + blaOXA-10 + blaAIM were detected in 2, 1 and 1 isolate(s), respectively. Five plasmid preparations harboring blaVEB-1 and blaOXA-10 were successfully transformed into chemically competent Escherichia coli DH5α with transformation efficiencies ranging between 6.8 × 10 3 and 3.7 × 10 4 CFU/μg DNA plasmid. Selected tested transformants were ceftazidime-resistant and harbored plasmids carrying blaOXA-10.
Conclusions: The study highlights the importance of the expeditious characterization of ESBLs and MBLs using genotypic methods among P. aeruginosa clinical isolates to hinder the development and dissemination of multidrug resistant strains.
Keywords: bla OXA-10; bla VIM; Curing; Genotype combinations; Transformation.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Conflict of interest: The authors declare that they have no competing interests. Ethics approval and consent to participate: The study was carried out in accordance with the guidelines and regulations of the Institutional Review Board (IRB) Committee, Faculty of Medicine, Alexandria University (IRB number: 00012098-FWA number: 00018699, serial number: 0306097). Consent to participate is not applicable for this study because this study does not contain any experiments with human participants and animals and the clinical isolates included in this study were obtained from existing clinical collections routinely assembled as part of laboratory practices of the medical microbiology lab at Alexandria Main University Hospital, Alexandria, Egypt. The need of consents was waived as neither the diagnosis nor the treatment was altered. Moreover, the data of the patients were not exposed. Consent for publication: Not applicable.
Figures



Similar articles
-
Emergence of blaVEB and blaGES among VIM-producing Pseudomonas aeruginosa clinical isolates in Alexandria, Egypt.Acta Microbiol Immunol Hung. 2019 Mar 1;66(1):131-142. doi: 10.1556/030.65.2018.044. Epub 2018 Nov 7. Acta Microbiol Immunol Hung. 2019. PMID: 30403360
-
Spread of extended-spectrum β-lactamase genes of blaOXA-10, blaPER-1 and blaCTX-M in Pseudomonas aeruginosa strains isolated from burn patients.Burns. 2014 Dec;40(8):1575-80. doi: 10.1016/j.burns.2014.02.008. Epub 2014 Apr 22. Burns. 2014. PMID: 24767142
-
Determination of extended spectrum beta-lactamases, metallo-beta-lactamases and AmpC-beta-lactamases among carbapenem resistant Pseudomonas aeruginosa isolated from burn patients.Burns. 2014 Dec;40(8):1556-61. doi: 10.1016/j.burns.2014.02.010. Epub 2014 Apr 22. Burns. 2014. PMID: 24767143
-
Characterization of antibiotic resistance profiles in Pseudomonas aeruginosa isolates from burn patients.Burns. 2021 Dec;47(8):1833-1843. doi: 10.1016/j.burns.2021.03.005. Epub 2021 Mar 17. Burns. 2021. PMID: 33795157 Free PMC article.
-
Current trends in the epidemiology of multidrug-resistant and beta-lactamase-producing Pseudomonas aeruginosa in Asia and Africa: a systematic review and meta-analysis.PeerJ. 2025 Feb 24;13:e18986. doi: 10.7717/peerj.18986. eCollection 2025. PeerJ. 2025. PMID: 40017659 Free PMC article.
Cited by
-
Advancements in Immunology and Microbiology Research: A Comprehensive Exploration of Key Areas.Microorganisms. 2024 Aug 14;12(8):1672. doi: 10.3390/microorganisms12081672. Microorganisms. 2024. PMID: 39203514 Free PMC article. Review.
-
Prevalence, trends, and molecular insights into colistin resistance among gram-negative bacteria in Egypt: a systematic review and meta-analysis.Ann Clin Microbiol Antimicrob. 2025 May 10;24(1):32. doi: 10.1186/s12941-025-00799-3. Ann Clin Microbiol Antimicrob. 2025. PMID: 40349047 Free PMC article.
References
-
- Peterson LR. Bad bugs, no drugs: no ESCAPE revisited. Clin Infect Dis. 2009;49:992–3. - PubMed
-
- Driscoll JA, Brody SL, Kollef MH. The epidemiology, pathogenesis and treatment of Pseudomonas aeruginosa infections. Drugs. 2007;67:351–68. - PubMed
-
- Gellatly SL, Hancock RE. Pseudomonas aeruginosa: new insights into pathogenesis and host defenses. Pathog Dis. 2013;67:159–73. - PubMed
-
- Chatterjee M, Anju C, Biswas L, Kumar VA, Mohan CG, Biswas R. Antibiotic resistance in Pseudomonas aeruginosa and alternative therapeutic options. Int J Med Microbiol. 2016;306:48–58. - PubMed
-
- Chaudhary M, Payasi A. Rising antimicrobial resistance of Pseudomonas aeruginosa isolated from clinical specimens in India. J Proteomics Bioinform. 2013;6:005–9.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

