Protocol for generating three-dimensional induced early ameloblasts using serum-free media and growth factors
- PMID: 38824640
- PMCID: PMC11176825
- DOI: 10.1016/j.xpro.2024.103100
Protocol for generating three-dimensional induced early ameloblasts using serum-free media and growth factors
Abstract
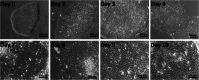
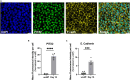
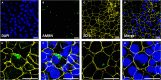
Adult humans cannot regenerate the enamel-forming cell type, ameloblasts. Hence, human induced pluripotent stem cell (hiPSC)-derived ameloblasts are valuable for investigating tooth development and regeneration. Here, we present a protocol for generating three-dimensional induced early ameloblasts (ieAMs) utilizing serum-free media and growth factors. We describe steps for directing hiPSCs toward oral epithelium and then toward ameloblast fate. These cells can form suspended early ameloblast organoids. This approach is critical for understanding, treating, and promoting regeneration in diseases like amelogenesis imperfecta. For complete details on the use and execution of this protocol, please refer to Alghadeer et al.1.
Keywords: Cell Differentiation; Developmental biology; Organoids; Stem Cells.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests A.A., A.P.P., J.M., and H.R.-B. are co-inventors on a patent application entitled “A Method to Direct the Differentiation of Human Induced Pluripotent Stem Cells into Early Ameloblasts” (PCT/US2022/053517 filed 12/20/2022).
Figures









References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

