Remodelin delays non-small cell lung cancer progression by inhibiting NAT10 via the EMT pathway
- PMID: 38826095
- PMCID: PMC11145023
- DOI: 10.1002/cam4.7283
Remodelin delays non-small cell lung cancer progression by inhibiting NAT10 via the EMT pathway
Abstract
Background: Lung cancer remains the foremost reason of cancer-related mortality, with invasion and metastasis profoundly influencing patient prognosis. N-acetyltransferase 10 (NAT10) catalyzes the exclusive N (4)-acetylcytidine (ac4C) modification in eukaryotic RNA. NAT10 dysregulation is linked to various diseases, yet its role in non-small cell lung cancer (NSCLC) invasion and metastasis remains unclear. Our study delves into the clinical significance and functional aspects of NAT10 in NSCLC.
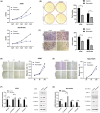
Methods: We investigated NAT10's clinical relevance using The Cancer Genome Atlas (TCGA) and a group of 98 NSCLC patients. Employing WB, qRT-PCR, and IHC analyses, we assessed NAT10 expression in NSCLC tissues, bronchial epithelial cells (BECs), NSCLC cell lines, and mouse xenografts. Further, knockdown and overexpression techniques (siRNA, shRNA, and plasmid) were employed to evaluate NAT10's effects. A series of assays were carried out, including CCK-8, colony formation, wound healing, and transwell assays, to elucidate NAT10's role in proliferation, invasion, and metastasis. Additionally, we utilized lung cancer patient-derived 3D organoids, mouse xenograft models, and Remodelin (NAT10 inhibitor) to corroborate these findings.
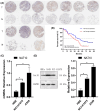
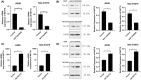
Results: Our investigations revealed high NAT10 expression in NSCLC tissues, cell lines and mouse xenograft models. High NAT10 level correlated with advanced T stage, lymph node metastasis and poor overall survive. NAT10 knockdown curtailed proliferation, invasion, and migration, whereas NAT10 overexpression yielded contrary effects. Furthermore, diminished NAT10 levels correlated with increased E-cadherin level whereas decreased N-cadherin and vimentin expressions, while heightened NAT10 expression displayed contrasting results. Notably, Remodelin efficiently attenuated NSCLC proliferation, invasion, and migration by inhibiting NAT10 through the epithelial-mesenchymal transition (EMT) pathway.
Conclusions: Our data underscore NAT10 as a potential therapeutic target for NSCLC, presenting avenues for targeted intervention against lung cancer through NAT10 inhibition.
Keywords: N‐acetyltransferase 10; Remodelin; epithelial‐to‐mesenchymal transition; invasion and metastasis; lung cancer; mouse xenograft; patients‐derived organoids; proliferation; tumorigenicity.
© 2024 The Author(s). Cancer Medicine published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors have no conflicts of interest to declare.
Figures




Similar articles
-
Emerging Role of NAT10 as ac4C Writer in Inflammatory Diseases: Mechanisms and Therapeutic Applications.Curr Drug Targets. 2025;26(4):282-294. doi: 10.2174/0113894501346709241202110834. Curr Drug Targets. 2025. PMID: 39633518 Review.
-
Acetyltransferase NAT10 promotes an immunosuppressive microenvironment by modulating CD8+ T cell activity in prostate cancer.Mol Biomed. 2024 Dec 9;5(1):67. doi: 10.1186/s43556-024-00228-5. Mol Biomed. 2024. PMID: 39648231 Free PMC article.
-
NAT10 Overexpression Promotes Tumorigenesis and Epithelial-Mesenchymal Transition Through AKT Pathway in Gastric Cancer.Dig Dis Sci. 2024 Sep;69(9):3261-3275. doi: 10.1007/s10620-024-08472-z. Epub 2024 Jul 11. Dig Dis Sci. 2024. PMID: 38990269
-
Targeting N4-acetylcytidine suppresses hepatocellular carcinoma progression by repressing eEF2-mediated HMGB2 mRNA translation.Cancer Commun (Lond). 2024 Sep;44(9):1018-1041. doi: 10.1002/cac2.12595. Epub 2024 Jul 19. Cancer Commun (Lond). 2024. PMID: 39030964 Free PMC article.
-
A Review of Research on the Mechanism of Tumor Regulation by N-Acetyltransferase 10.Discov Med. 2024 Jul;36(186):1334-1344. doi: 10.24976/Discov.Med.202436186.123. Discov Med. 2024. PMID: 39054704 Review.
Cited by
-
Emerging Role of NAT10 as ac4C Writer in Inflammatory Diseases: Mechanisms and Therapeutic Applications.Curr Drug Targets. 2025;26(4):282-294. doi: 10.2174/0113894501346709241202110834. Curr Drug Targets. 2025. PMID: 39633518 Review.
-
RNA N4-acetylcytidine modification and its role in health and diseases.MedComm (2020). 2025 Jan 3;6(1):e70015. doi: 10.1002/mco2.70015. eCollection 2025 Jan. MedComm (2020). 2025. PMID: 39764566 Free PMC article. Review.
-
NAT10 promotes radiotherapy resistance in non-small cell lung cancer by regulating KPNB1-mediated PD-L1 nuclear translocation.Open Life Sci. 2025 Mar 18;20(1):20251065. doi: 10.1515/biol-2025-1065. eCollection 2025. Open Life Sci. 2025. PMID: 40109769 Free PMC article.
-
NAT10 Knockdown Improves Cisplatin Sensitivity in Non-Small Cell Lung Cancer by Inhibiting the TRIM44/PI3K/AKT Pathway.Thorac Cancer. 2025 May;16(9):e70079. doi: 10.1111/1759-7714.70079. Thorac Cancer. 2025. PMID: 40324967 Free PMC article.
-
Role of NAT10-mediated ac4C acetylation of ENO1 mRNA in glycolysis and apoptosis in non-small cell lung cancer cells.BMC Pulm Med. 2025 Feb 13;25(1):75. doi: 10.1186/s12890-024-03463-2. BMC Pulm Med. 2025. PMID: 39948547 Free PMC article.
References
-
- Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72(1):7‐33. - PubMed
-
- Gridelli C, Rossi A, Carbone DP, et al. Non‐small‐cell lung cancer. Nat Rev Dis Primers. 2015;1:15009. - PubMed
-
- Herbst RS, Morgensztern D, Boshoff C. The biology and management of non‐small cell lung cancer. Nature. 2018;553(7689):446‐454. - PubMed
-
- Arbour KC, Riely GJ. Systemic therapy for locally advanced and metastatic non–small cell lung cancer: a review. JAMA. 2019;322(8):764‐774. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

