Cryptic diversity and speciation in an endemic copepod crustacean Harpacticella inopinata within Lake Baikal
- PMID: 38826165
- PMCID: PMC11140236
- DOI: 10.1002/ece3.11471
Cryptic diversity and speciation in an endemic copepod crustacean Harpacticella inopinata within Lake Baikal
Abstract
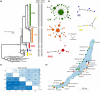
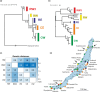
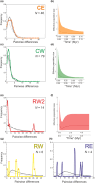
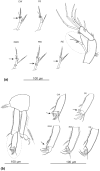
Ancient lakes are hotspots of species diversity, posing challenges and opportunities for exploration of the dynamics of endemic diversification. Lake Baikal in Siberia, the oldest lake in the world, hosts a particularly rich crustacean fauna, including the largest known species flock of harpacticoid copepods with some 70 species. Here, we focused on exploring the diversity and evolution within a single nominal species, Harpacticella inopinata Sars, 1908, using molecular markers (mitochondrial COI, nuclear ITS1 and 28S rRNA) and a set of qualitative and quantitative morphological traits. Five major mitochondrial lineages were recognized, with model-corrected COI distances of 0.20-0.37. A concordant pattern was seen in the nuclear data set, and qualitative morphological traits also distinguish a part of the lineages. All this suggests the presence of several hitherto unrecognized cryptic taxa within the baikalian H. inopinata, with long independent histories. The abundances, distributions and inferred demographic histories were different among taxa. Two taxa, H. inopinata CE and H. inopinata CW, were widespread on the eastern and western coasts, respectively, and were largely allopatric. Patterns in mitochondrial variation, that is, shallow star-like haplotype networks, suggest these taxa have spread through the lake relatively recently. Three other taxa, H. inopinata RE, RW and RW2, instead were rare and had more localized distributions on either coast, but showed deeper intraspecies genealogies, suggesting older regional presence. The rare taxa were often found in sympatry with the others and occasionally introgressed by mtDNA from the common ones. The mitochondrial divergence between and within the H. inopinata lineages is still unexpectedly deep, suggesting an unusually high molecular rate. The recognition of true systematic diversity in the evaluation and management of ecosystems is important in hotspots, as it is everywhere else, while the translation of the diversity into a formal taxonomy remains a challenge.
Keywords: Copepoda; ancient lakes; endemism; phylogeography; speciation.
© 2024 The Authors. Ecology and Evolution published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
The endemic Cladophorales (Ulvophyceae) of ancient Lake Baikal represent a monophyletic group of very closely related but morphologically diverse species.J Phycol. 2018 Oct;54(5):616-629. doi: 10.1111/jpy.12773. Epub 2018 Sep 12. J Phycol. 2018. PMID: 30076711
-
Cryptic diversity in the Western Balkan endemic copepod: Four species in one?Mol Phylogenet Evol. 2016 Jul;100:124-134. doi: 10.1016/j.ympev.2016.04.010. Epub 2016 Apr 7. Mol Phylogenet Evol. 2016. PMID: 27063254
-
Morphological and molecular diversity of Lake Baikal candonid ostracods, with description of a new genus.Zookeys. 2017 Jul 11;(684):19-56. doi: 10.3897/zookeys.684.13249. eCollection 2017. Zookeys. 2017. PMID: 28769732 Free PMC article.
-
Biogeography and speciation of terrestrial fauna in the south-western Australian biodiversity hotspot.Biol Rev Camb Philos Soc. 2015 Aug;90(3):762-93. doi: 10.1111/brv.12132. Epub 2014 Aug 15. Biol Rev Camb Philos Soc. 2015. PMID: 25125282 Review.
-
The taxonomic diversity of the cichlid fish fauna of ancient Lake Tanganyika, East Africa.J Great Lakes Res. 2020 Oct;46(5):1067-1078. doi: 10.1016/j.jglr.2019.05.009. J Great Lakes Res. 2020. PMID: 33100489 Free PMC article. Review.
Cited by
-
An integrative approach to the delimitation of pseudocryptic species in the Eucyclopssperatus complex (Copepoda, Cyclopoida) with a description of a new species.Zookeys. 2025 Feb 7;1226:217-260. doi: 10.3897/zookeys.1226.138389. eCollection 2025. Zookeys. 2025. PMID: 39959868 Free PMC article.
-
Rediscovered historical lots uncover the identity of Nikolaj A. Livanow's Protoclepsis species and clarify the taxonomy of Palearctic waterfowl-associated leeches (Hirudinea: Glossiphoniidae).Syst Parasitol. 2025 Apr 25;102(3):32. doi: 10.1007/s11230-025-10221-3. Syst Parasitol. 2025. PMID: 40278972
References
-
- Baek, S. Y. , Jang, K. H. , Choi, E. H. , Ryu, S. H. , Kim, S. K. , Lee, J. H. , Lim, Y. J. , Lee, J. , Jun, J. , Kwak, M. , Lee, Y. S. , Hwang, J. S. , Maran, B. A. V. , Chang, C. Y. , Kim, I. H. , & Hwang, U. W. (2016). DNA barcoding of metazoan zooplankton copepods from South Korea. PLoS ONE, 11(7), e0157307. 10.1371/journal.pone.0157307 - DOI - PMC - PubMed
-
- Barluenga, M. , Stolting, K. N. , Salzburger, W. , Muschick, M. , & Meyer, A. (2006). Sympatric speciation in Nicaraguan crater lake cichlid fish. Nature, 439, 719–723. - PubMed
-
- Barreto, F. S. , Watson, E. T. , Lima, T. G. , Willett, C. S. , Edmands, S. , Li, W. , & Burton, R. S. (2018). Genomic signatures of mitonuclear coevolution across populations of Tigriopus californicus . Nature Ecology & Evolution, 2018(2), 1250–1257. - PubMed
-
- Bielby, J. , Cardillo, M. , Cooper, N. , & Purvis, A. (2010). Modelling extinction risk in multispecies data sets: Phylogenetically independent contrasts versus decision trees. Biodiversity and Conservation, 19(1), 113–127.
Associated data
LinkOut - more resources
Full Text Sources
Miscellaneous

