This is a preprint.
Single-molecule imaging reveals the kinetics of non-homologous end-joining in living cells
- PMID: 38826211
- PMCID: PMC11142080
- DOI: 10.1101/2023.06.22.546088
Single-molecule imaging reveals the kinetics of non-homologous end-joining in living cells
Update in
-
Single-molecule imaging reveals the kinetics of non-homologous end-joining in living cells.Nat Commun. 2024 Nov 23;15(1):10159. doi: 10.1038/s41467-024-54545-y. Nat Commun. 2024. PMID: 39578493 Free PMC article.
Abstract
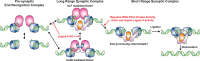
Non-homologous end joining (NHEJ) is the predominant pathway that repairs DNA double-stranded breaks (DSBs) in vertebrates. However, due to challenges in detecting DSBs in living cells, the repair capacity of the NHEJ pathway is unknown. The DNA termini of many DSBs must be processed to allow ligation while minimizing genetic changes that result from break repair. Emerging models propose that DNA termini are first synapsed ~115Å apart in one of several long-range synaptic complexes before transitioning into a short-range synaptic complex that juxtaposes DNA ends to facilitate ligation. The transition from long-range to short-range synaptic complexes involves both conformational and compositional changes of the NHEJ factors bound to the DNA break. Importantly, it is unclear how NHEJ proceeds in vivo because of the challenges involved in analyzing recruitment of NHEJ factors to DSBs over time in living cells. Here, we develop a new approach to study the temporal and compositional dynamics of NHEJ complexes using live cell single-molecule imaging. Our results provide direct evidence for stepwise maturation of the NHEJ complex, pinpoint key regulatory steps in NHEJ progression, and define the overall repair capacity NHEJ in living cells.
Conflict of interest statement
COMPETING INTERESTS None declared.
Figures






Similar articles
-
Single-molecule imaging reveals the kinetics of non-homologous end-joining in living cells.Nat Commun. 2024 Nov 23;15(1):10159. doi: 10.1038/s41467-024-54545-y. Nat Commun. 2024. PMID: 39578493 Free PMC article.
-
Analysis of chromatid-break-repair detects a homologous recombination to non-homologous end-joining switch with increasing load of DNA double-strand breaks.Mutat Res Genet Toxicol Environ Mutagen. 2021 Jul;867:503372. doi: 10.1016/j.mrgentox.2021.503372. Epub 2021 Jun 12. Mutat Res Genet Toxicol Environ Mutagen. 2021. PMID: 34266628
-
Holding it together: DNA end synapsis during non-homologous end joining.DNA Repair (Amst). 2023 Oct;130:103553. doi: 10.1016/j.dnarep.2023.103553. Epub 2023 Aug 8. DNA Repair (Amst). 2023. PMID: 37572577 Free PMC article. Review.
-
Mechanisms of DNA double strand break repair and chromosome aberration formation.Cytogenet Genome Res. 2004;104(1-4):14-20. doi: 10.1159/000077461. Cytogenet Genome Res. 2004. PMID: 15162010 Review.
-
Tousled-like kinase loss confers PARP inhibitor resistance in BRCA1-mutated cancers by impeding non-homologous end joining repair.Mol Med. 2025 Jan 22;31(1):18. doi: 10.1186/s10020-025-01066-z. Mol Med. 2025. PMID: 39844055 Free PMC article.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
