This is a preprint.
Single-cell transcriptomics reveals stage- and side-specificity of gene modules in colorectal cancer
- PMID: 38826219
- PMCID: PMC11142301
- DOI: 10.21203/rs.3.rs-4402565/v1
Single-cell transcriptomics reveals stage- and side-specificity of gene modules in colorectal cancer
Abstract
Background: An understanding of mechanisms underlying colorectal cancer (CRC) development and progression is yet to be fully elucidated. This study aims to employ network theoretic approaches to analyse single cell transcriptomic data from CRC to better characterize its progression and sided-ness.
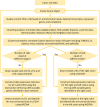
Methods: We utilized a recently published single-cell RNA sequencing data (GEO-GSE178341) and parsed the cell X gene data by stage and side (right and left colon). Using Weighted Gene Co-expression Network Analysis (WGCNA), we identified gene modules with varying preservation levels (weak or strong) of network topology between early (pT1) and late stages (pT234), and between right and left colons. Spearman's rank correlation (ρ) was used to assess the similarity or dissimilarity in gene connectivity.
Results: Equalizing cell counts across different stages, we detected 13 modules for the early stage, two of which were non-preserved in late stages. Both non-preserved modules displayed distinct gene connectivity patterns between the early and late stages, characterized by low ρ values. One module predominately dealt with myeloid cells, with genes mostly enriched for cytokine-cytokine receptor interaction potentiallystimulating myeloid cells to participate in angiogenesis. The second module, representing a subset of epithelial cells, was mainly enriched for carbohydrate digestion and absorption, influencing the gut microenvironment through the breakdown of carbohydrates. In the comparison of left vs. right colons, two of 12 modules identified in the right colon were non-preserved in the left colon. One captured a small fraction of epithelial cells and was enriched for transcriptional misregulation in cancer, potentially impacting communication between epithelial cells and the tumor microenvironment. The other predominantly contained B cells with a crucial role in maintaining human gastrointestinal health and was enriched for B-cell receptor signalling pathway.
Conclusions: We identified modules with topological and functional differences specific to cell types between the early and late stages, and between the right and left colons. This study enhances the understanding of roles played by different cell types at different stages and sides, providing valuable insights for future studies focused on the diagnosis and treatment of CRC.
Keywords: WGCNA; early tumor stage; functional enrichment; late tumor stage; left colon; modularity; right colon; scRNA-seq.
Conflict of interest statement
Competing interests The authors declare that they have no competing interests.
Figures



Similar articles
-
Holistic exploration of CHGA and hsa-miR-137 in colorectal cancer via multi-omic data Integration.Heliyon. 2024 Mar 3;10(5):e27046. doi: 10.1016/j.heliyon.2024.e27046. eCollection 2024 Mar 15. Heliyon. 2024. PMID: 38495181 Free PMC article.
-
Metabolic Heterogeneity of Tumor Cells and its Impact on Colon Cancer Metastasis: Insights from Single-Cell and Bulk Transcriptome Analyses.J Cancer. 2024 Jun 3;15(13):4175-4196. doi: 10.7150/jca.94630. eCollection 2024. J Cancer. 2024. PMID: 38947396 Free PMC article.
-
Coding and non-coding co-expression network analysis identifies key modules and driver genes associated with precursor lesions of gastric cancer.Genomics. 2022 May;114(3):110370. doi: 10.1016/j.ygeno.2022.110370. Epub 2022 Apr 14. Genomics. 2022. PMID: 35430283
-
The common transcriptional subnetworks of the grape berry skin in the late stages of ripening.BMC Plant Biol. 2017 May 30;17(1):94. doi: 10.1186/s12870-017-1043-1. BMC Plant Biol. 2017. PMID: 28558655 Free PMC article.
-
Identifying miRNA and gene modules of colon cancer associated with pathological stage by weighted gene co-expression network analysis.Onco Targets Ther. 2018 May 15;11:2815-2830. doi: 10.2147/OTT.S163891. eCollection 2018. Onco Targets Ther. 2018. PMID: 29844680 Free PMC article.
References
-
- Siegel RL, Miller KD, Wagle NS, Jemal A: Cancer statistics, 2023. CA: a cancer journal for clinicians 2023, 73(1):17–48. - PubMed
-
- Brenner H, Kloor M, Pox CP: Colorectal cancer. Lancet 2014, 383(9927):1490–1502. - PubMed
-
- Amin MB, Greene FL, Edge SB, Compton CC, Gershenwald JE, Brookland RK, Meyer L, Gress DM, Byrd DR, Winchester DP: The Eighth Edition AJCC Cancer Staging Manual: Continuing to build a bridge from a population-based to a more “personalized” approach to cancer staging. CA: a cancer journal for clinicians 2017, 67(2):93–99. - PubMed
-
- Hanna DL, Lenz HJ: How We Treat Left-Sided vs Right-Sided Colon Cancer. Clin Adv Hematol Onc 2020, 18(5):253–257. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

