This is a preprint.
Microfluidic Ecology Unravels the Genetic and Ecological Drivers of T4r Bacteriophage Resistance in E. coli: Insights into Biofilm-Mediated Evolution
- PMID: 38826273
- PMCID: PMC11142369
- DOI: 10.21203/rs.3.rs-4356333/v1
Microfluidic Ecology Unravels the Genetic and Ecological Drivers of T4r Bacteriophage Resistance in E. coli: Insights into Biofilm-Mediated Evolution
Abstract
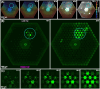
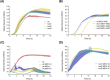
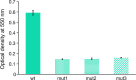
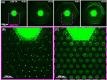
We use a microfluidic ecology which generates non-uniform phage concentration gradients and micro-ecological niches to reveal the importance of time, spatial population structure and collective population dynamics in the de novo evolution of T4r bacteriophage resistant motile E. coli. An insensitive bacterial population against T4r phage occurs within 20 hours in small interconnected population niches created by a gradient of phage virions, driven by evolution in transient biofilm patches. Sequencing of the resistant bacteria reveals mutations at the receptor site of bacteriophage T4r as expected but also in genes associated with biofilm formation and surface adhesion, supporting the hypothesis that evolution within transient biofilms drives de novo phage resistance.
Conflict of interest statement
Additional Declarations: There is NO Competing Interest.
Figures


Similar articles
-
[Biological characteristics and genomic information of a bacteriophage against pan-drug resistant Klebsiella pneumoniae in a burn patient and its effects on bacterial biofilm].Zhonghua Shao Shang Za Zhi. 2020 Jan 20;36(1):14-23. doi: 10.3760/cma.j.issn.1009-2587.2020.01.004. Zhonghua Shao Shang Za Zhi. 2020. PMID: 32023713 Chinese.
-
Ecology of Anti-Biofilm Agents II: Bacteriophage Exploitation and Biocontrol of Biofilm Bacteria.Pharmaceuticals (Basel). 2015 Sep 9;8(3):559-89. doi: 10.3390/ph8030559. Pharmaceuticals (Basel). 2015. PMID: 26371011 Free PMC article. Review.
-
Improving Phage-Biofilm In Vitro Experimentation.Viruses. 2021 Jun 19;13(6):1175. doi: 10.3390/v13061175. Viruses. 2021. PMID: 34205417 Free PMC article.
-
Biofilm Structure Promotes Coexistence of Phage-Resistant and Phage-Susceptible Bacteria.mSystems. 2020 Jun 23;5(3):e00877-19. doi: 10.1128/mSystems.00877-19. mSystems. 2020. PMID: 32576653 Free PMC article.
-
Big Impact of the Tiny: Bacteriophage-Bacteria Interactions in Biofilms.Trends Microbiol. 2019 Sep;27(9):739-752. doi: 10.1016/j.tim.2019.04.006. Epub 2019 May 22. Trends Microbiol. 2019. PMID: 31128928 Review.
References
-
- Pires D. P., Melo L. D. R., and Azeredo J.. Understanding the complex phage-host interactions in biofilm communities. Annu Rev Virol, 8(1):73–94, 2021. - PubMed
-
- Jablonka E. and Raz G.. Transgenerational epigenetic inheritance: Prevalence, mechanisms, and implications for the study of heredity and evolution. Quarterly Review of Biology, 84(2):131–176, 2009. 446lx Times Cited:935 Cited References Count:236. - PubMed
Publication types
LinkOut - more resources
Full Text Sources

