This is a preprint.
A PARP2-specific active site α-helix melts to permit DNA damage-induced enzymatic activation
- PMID: 38826291
- PMCID: PMC11142140
- DOI: 10.1101/2024.05.20.594972
A PARP2-specific active site α-helix melts to permit DNA damage-induced enzymatic activation
Update in
-
A PARP2 active site helix melts to permit DNA damage-induced enzymatic activation.Mol Cell. 2025 Mar 6;85(5):865-876.e4. doi: 10.1016/j.molcel.2025.01.004. Epub 2025 Jan 30. Mol Cell. 2025. PMID: 39889708
Abstract
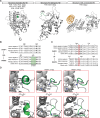
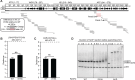
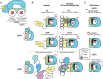
PARP1 and PARP2 recognize DNA breaks immediately upon their formation, generate a burst of local PARylation to signal their location, and are co-targeted by all current FDA-approved forms of PARP inhibitors (PARPi) used in the cancer clinic. Recent evidence indicates that the same PARPi molecules impact PARP2 differently from PARP1, raising the possibility that allosteric activation may also differ. We find that unlike for PARP1, destabilization of the autoinhibitory domain of PARP2 is insufficient for DNA damage-induced catalytic activation. Rather, PARP2 activation requires further unfolding of an active site α-helix absent in PARP1. Only one clinical PARPi, Olaparib, stabilizes the PARP2 active site α-helix, representing a structural feature with the potential to discriminate small molecule inhibitors. Collectively, our findings reveal unanticipated differences in local structure and changes in activation-coupled backbone dynamics between PARP1 and PARP2.
Conflict of interest statement
Declarations of interest B.E.B., J.M.P., and T.T.T. are co-founders of Hysplex, Inc. with interests in PARP inhibitor development. B.E.B., J.M.P., and T.T.T. are co-inventors on a provisional patent application filed by UPenn that is related to this work. B.E.B is on the scientific advisory board of Denovicon Therapeutics.
Figures



Similar articles
-
A PARP2 active site helix melts to permit DNA damage-induced enzymatic activation.Mol Cell. 2025 Mar 6;85(5):865-876.e4. doi: 10.1016/j.molcel.2025.01.004. Epub 2025 Jan 30. Mol Cell. 2025. PMID: 39889708
-
Clinical PARP inhibitors allosterically induce PARP2 retention on DNA.Sci Adv. 2023 Mar 24;9(12):eadf7175. doi: 10.1126/sciadv.adf7175. Epub 2023 Mar 24. Sci Adv. 2023. PMID: 36961901 Free PMC article.
-
PARP inhibitors trap PARP2 and alter the mode of recruitment of PARP2 at DNA damage sites.Nucleic Acids Res. 2022 Apr 22;50(7):3958-3973. doi: 10.1093/nar/gkac188. Nucleic Acids Res. 2022. PMID: 35349716 Free PMC article.
-
DNA Repair Enzyme Poly(ADP-Ribose) Polymerase 1/2 (PARP1/2)-Targeted Nuclear Imaging and Radiotherapy.Cancers (Basel). 2022 Feb 23;14(5):1129. doi: 10.3390/cancers14051129. Cancers (Basel). 2022. PMID: 35267438 Free PMC article. Review.
-
Medicinal chemistry approaches of poly ADP-Ribose polymerase 1 (PARP1) inhibitors as anticancer agents - A recent update.Eur J Med Chem. 2019 Mar 1;165:198-215. doi: 10.1016/j.ejmech.2019.01.024. Epub 2019 Jan 12. Eur J Med Chem. 2019. PMID: 30684797 Review.
References
Publication types
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
