This is a preprint.
Inducible transposon mutagenesis for genome-scale forward genetics
- PMID: 38826325
- PMCID: PMC11142078
- DOI: 10.1101/2024.05.21.595064
Inducible transposon mutagenesis for genome-scale forward genetics
Update in
-
Inducible transposon mutagenesis identifies bacterial fitness determinants during infection in mice.Nat Microbiol. 2025 May;10(5):1171-1183. doi: 10.1038/s41564-025-01975-z. Epub 2025 Mar 27. Nat Microbiol. 2025. PMID: 40148565 Free PMC article.
Abstract
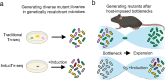
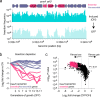
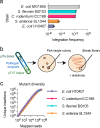
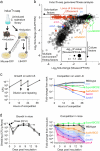
Transposon insertion sequencing (Tn-seq) is a powerful method for genome-scale functional genetics in bacteria. However, its effectiveness is often limited by a lack of mutant diversity, caused by either inefficient transposon delivery or stochastic loss of mutants due to population bottlenecks. Here, we introduce "InducTn-seq", which leverages inducible mutagenesis for temporal control of transposition. InducTn-seq generates millions of transposon mutants from a single colony, enabling the sensitive detection of subtle fitness defects and transforming binary classifications of gene essentiality into a quantitative fitness measurement across both essential and non-essential genes. Using a mouse model of infectious colitis, we show that InducTn-seq bypasses a highly restrictive host bottleneck to generate a diverse transposon mutant population from the few cells that initiate infection, revealing the role of oxygen-related metabolic plasticity in pathogenesis. Overall, InducTn-seq overcomes the limitations of traditional Tn-seq, unlocking new possibilities for genome-scale forward genetic screens in bacteria.
Conflict of interest statement
Competing interests The authors declare no competing interests.
Figures


References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
