This is a preprint.
Parallel evolution of X chromosome-specific SMC complexes in two nematode lineages
- PMID: 38826443
- PMCID: PMC11142195
- DOI: 10.1101/2024.05.21.595224
Parallel evolution of X chromosome-specific SMC complexes in two nematode lineages
Abstract
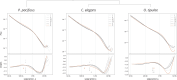
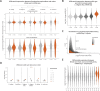
Mechanisms of X chromosome dosage compensation have been studied extensively in three model organisms that represent distinct clades. The diversity within each clade as a function of sex chromosome evolution though is largely unknown. Here, we anchor ourselves to the nematode Caenorhabditis elegans, where dosage compensation is accomplished by an X chromosome specific condensin that belongs to the family of structural maintenance of chromosomes (SMC) complexes. By combining a phylogenetic analyses of the C. elegans dosage compensation complex with a comparative analysis of its epigenetic signatures, such as X-specific topologically associating domains (TADs) and enrichment of H4K20me1, we show that the condensin-mediated mechanism evolved recently in the lineage leading to Caenorhabditis following an SMC-4 duplication. Unexpectedly, we found an independent duplication of SMC-4 in Pristionchus pacificus along with the presence of X-specific TADs and H4K20me1 enrichment, which suggests that condensin-mediated dosage compensation evolved more than once in nematodes. Differential expression analysis between sexes in several nematode species indicates that dosage compensation itself precedes the evolution of X-specific condensins. In Rhabditina, X-specific condensins may have evolved in the presence of an existing mechanism linked to H4K20 methylation as Oscheius tipulae X chromosomes are enriched for H4K20me1 without SMC-4 duplication or TADs. In contrast, Steinernema hermaphroditum lacks H4K20me1 enrichment, SMC-4 duplication, and TADs. Together, our results indicate that dosage compensation mechanisms continue to evolve in species with shared X chromosome ancestry, and SMC complexes may have been coopted repeatedly in nematodes, suggesting that the process of evolving chromosome wide gene regulatory mechanisms are constrained.
Keywords: H4K20me1; Hi-C; SMC complexes; Structural Maintenance of Chromosomes; TAD; X chromosome; caenorhabditis; condensin; dosage compensation; evolution; nematodes; oscheius; parallel evolution; pristionchus; sex chromosomes; steinernema; transcription.
Conflict of interest statement
CONFLICTING INTERESTS None.
Figures



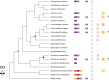
 , nigon element;
, nigon element;  , dosage compensation;
, dosage compensation;  , X specific TADS;
, X specific TADS;  no TADs on X chromosome;
no TADs on X chromosome;  , X specific enrichment of H4K20me1.
, X specific enrichment of H4K20me1.Similar articles
-
Short-Term Memory Impairment.2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 31424720 Free Books & Documents.
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
-
Systemic Inflammatory Response Syndrome.2025 Jun 20. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jun 20. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 31613449 Free Books & Documents.
-
A cross-species analysis of neuroanatomical covariance sex differences in humans and mice.Biol Sex Differ. 2025 Jul 1;16(1):47. doi: 10.1186/s13293-025-00728-1. Biol Sex Differ. 2025. PMID: 40598550 Free PMC article.
-
Carbamazepine versus phenytoin monotherapy for epilepsy: an individual participant data review.Cochrane Database Syst Rev. 2017 Feb 27;2(2):CD001911. doi: 10.1002/14651858.CD001911.pub3. Cochrane Database Syst Rev. 2017. Update in: Cochrane Database Syst Rev. 2019 Jul 18;7:CD001911. doi: 10.1002/14651858.CD001911.pub4. PMID: 28240353 Free PMC article. Updated.
References
-
- Charlesworth B. & Charlesworth D. A Model for the Evolution of Dioecy and Gynodioecy. The American Naturalist 112, 975–997 (1978).
-
- Hodgkin J. Genetic sex determination mechanisms and evolution. Bioessays 14, 253–261 (1992). - PubMed
-
- Charlesworth B. The evolution of chromosomal sex determination and dosage compensation. Curr Biol 6, 149–162 (1996). - PubMed
-
- Jablonka E. & Lamb M. J. The evolution of heteromorphic sex chromosomes. Biol Rev Camb Philos Soc 65, 249–276 (1990). - PubMed
-
- Charlesworth B. The evolution of sex chromosomes. Science 251, 1030–1033 (1991). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
