Exploring the Application of Metagenomic Next-Generation Sequencing in the Diagnosis of Unexplained Pulmonary Infection
- PMID: 38826507
- PMCID: PMC11141768
- DOI: 10.2147/IJGM.S459373
Exploring the Application of Metagenomic Next-Generation Sequencing in the Diagnosis of Unexplained Pulmonary Infection
Abstract
Background: Pulmonary infections are significant global health burdens, and conventional diagnostic methods (culture and polymerase chain reaction), are often limited by slow results and low sensitivity. Metagenomic next-generation sequencing (mNGS) offers a rapid, comprehensive alternative for identifying diverse pathogens, including rare and mixed infections. Thus, we assessed the diagnostic performance of mNGS in pulmonary infections, compared the findings with those of traditional pathogen detection methods, and explored its potential to enhance clinical diagnostics and patient care.
Methods: We collected samples from 125 immunocompromised patients diagnosed with pulmonary infection at the Department of Respiratory Medicine of Shenzhen Longgang Central Hospital from March 2020 to July 2022. We compared the rate of pathogen positivity and pathogen distribution between conventional pathogen detection methods and mNGS using samples including sputum, blood, and bronchoalveolar lavage fluid.
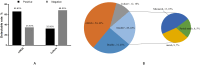
Results: Among the 125 cases of unexplained pulmonary infection, 82 (65.6%) and 40 (32.0%) tested positive for pathogens using mNGS and routine culture, respectively (P < 0.05). Both methods of pathogen detection were positive in 28 (22.4%) cases (complete match, 9; complete mismatch, 13; partial match, 6). However, 43.2% of cases only tested positive using mNGS, 9.4% only tested positive using routine tests, and 24.8% tested negative using both methods. A viral infection was present in 55.2% of cases. The detection rate of mycobacteria using mNGS (12.8%) was higher than that using conventional pathogen detection methods (5.6%).
Conclusion: mNGS technology enhances pathogen detection in unexplained pulmonary infections, enabling targeted antimicrobial therapy and consequently helping to reduce broad-spectrum antibiotic use, aligning treatments more closely with the causative pathogens. Thus, mNGS offers significant clinical value by improving treatment efficacy and potentially reducing antibiotic resistance in pulmonary infection cases.
Keywords: BALF; Pathogens; Unexplained pulmonary infection; mNGS.
© 2024 Chen et al.
Conflict of interest statement
The authors declare that they have no competing interests in this work.
Figures

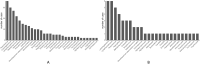

Similar articles
-
Improving pulmonary infection diagnosis with metagenomic next-generation sequencing of bronchoalveolar lavage fluid.J Med Microbiol. 2024 Feb;73(2). doi: 10.1099/jmm.0.001808. J Med Microbiol. 2024. PMID: 38420890
-
Significance of Combining Bronchoalveolar Lavage Fluid With Targeted Next-Generation Sequencing in the Pathogen Detection-Based Diagnosis of Pulmonary Infections.Clin Respir J. 2025 Jan;19(1):e70046. doi: 10.1111/crj.70046. Clin Respir J. 2025. PMID: 39835383 Free PMC article.
-
Improving Suspected Pulmonary Infection Diagnosis by Bronchoalveolar Lavage Fluid Metagenomic Next-Generation Sequencing: a Multicenter Retrospective Study.Microbiol Spectr. 2022 Aug 31;10(4):e0247321. doi: 10.1128/spectrum.02473-21. Epub 2022 Aug 9. Microbiol Spectr. 2022. PMID: 35943274 Free PMC article.
-
Diagnostic challenges in peritoneal dialysis-associated peritonitis with atypical and rare pathogens: A new era of metagenomic next-generation sequencing precision diagnosis.Perit Dial Int. 2025 May 19:8968608251333879. doi: 10.1177/08968608251333879. Online ahead of print. Perit Dial Int. 2025. PMID: 40388929 Review.
-
Clinical metagenomics-challenges and future prospects.Front Microbiol. 2023 Jun 28;14:1186424. doi: 10.3389/fmicb.2023.1186424. eCollection 2023. Front Microbiol. 2023. PMID: 37448579 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources

