Investigating the Taenia solium Fatty Acid Binding Protein Superfamily for Their Immunological Outlook and Prospect for Therapeutic Targets
- PMID: 38826528
- PMCID: PMC11137695
- DOI: 10.1021/acsomega.3c09253
Investigating the Taenia solium Fatty Acid Binding Protein Superfamily for Their Immunological Outlook and Prospect for Therapeutic Targets
Abstract
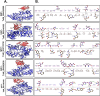
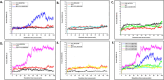
Taenia solium, like other helminthic parasites, lacks key components of cellular machinery required for endogenous lipid biosynthesis. This deficiency compels the parasite to obtain all of its lipid requirements from its host. The passage of lipids across the cell membrane is tightly regulated. To facilitate effective lipid transport, the cestode parasite utilizes certain lipid binding proteins called FABPs. These FABPs bind with the lipid ligands and allow the transport of lipids across the membranes and into the cytosol. Here, by integrating a computational with homology protein prediction tools, we had identified five FABPs in the T. solium proteome. We confirmed their presence by RNA expression analysis of respective genes from the parasite's cysticerci transcript. During the molecular modeling and MD simulation studies, two of them, TsM_000544100 and TsM_001185100, were most stable. Furthermore, they had a robust interaction with the IgG1 molecule, as evidenced by MD simulation. In addition, by employing in silico screening, we had identified potential ligand interacting residues that are present on the probable druggable site. In combination with in vitro cysticidal assays, enalaprilat dihydrate showed efficacy against cysticerci, which suggests that FABPs play a significant role in the cysticercus life cycle. Together, we provided a detailed distribution of all FABPs expressed by T. solium cysticerci and the critical role of TsM_001185100 in cysticercus viability.
© 2024 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures








Similar articles
-
Differential in vitro effects of insulin on Taenia crassiceps and Taenia solium cysticerci.J Helminthol. 2009 Dec;83(4):403-12. doi: 10.1017/S0022149X09990265. Epub 2009 Jun 24. J Helminthol. 2009. PMID: 19549345
-
A novel progesterone receptor membrane component (PGRMC) in the human and swine parasite Taenia solium: implications to the host-parasite relationship.Parasit Vectors. 2018 Mar 9;11(1):161. doi: 10.1186/s13071-018-2703-1. Parasit Vectors. 2018. PMID: 29523160 Free PMC article.
-
Cysticidal effect of a pure naphthoquinone on Taenia crassiceps cysticerci.Parasitol Res. 2021 Nov;120(11):3783-3794. doi: 10.1007/s00436-021-07281-x. Epub 2021 Sep 22. Parasitol Res. 2021. PMID: 34549347
-
Steroid hormone production by parasites: the case of Taenia crassiceps and Taenia solium cysticerci.J Steroid Biochem Mol Biol. 2003 Jun;85(2-5):221-5. doi: 10.1016/s0960-0760(03)00233-4. J Steroid Biochem Mol Biol. 2003. PMID: 12943707 Review.
-
Cytosolic fatty acid binding proteins catalyze two distinct steps in intracellular transport of their ligands.Mol Cell Biochem. 2002 Oct;239(1-2):35-43. Mol Cell Biochem. 2002. PMID: 12479566 Review.
References
-
- Arora N.; Tripathi S.; Sao R.; Mondal P.; Mishra A.; Prasad A. Molecular Neuro-Pathomechanism of Neurocysticercosis: How Host Genetic Factors Influence Disease Susceptibility. Mol. Neurobiol 2018, 55 (2), 1019–1025. 10.1007/s12035-016-0373-6. - DOI - PubMed
- Prasad K. N.; Prasad A.; Verma A.; Singh A. K. Human cysticercosis and Indian scenario: a review. J. Biosci 2008, 33 (4), 571–582. 10.1007/s12038-008-0075-y. - DOI - PubMed
- Singh A. K.; Prasad K. N.; Prasad A.; Tripathi M.; Gupta R. K.; Husain N. Immune responses to viable and degenerative metacestodes of Taenia solium in naturally infected swine. Int. J. Parasitol 2013, 43 (14), 1101–1107. 10.1016/j.ijpara.2013.07.009. - DOI - PubMed
LinkOut - more resources
Full Text Sources
