Microbiome in the nasopharynx: Insights into the impact of COVID-19 severity
- PMID: 38826746
- PMCID: PMC11141365
- DOI: 10.1016/j.heliyon.2024.e31562
Microbiome in the nasopharynx: Insights into the impact of COVID-19 severity
Abstract
Background: The respiratory tract harbors a variety of microbiota, whose composition and abundance depend on specific site factors, interaction with external factors, and disease. The aim of this study was to investigate the relationship between COVID-19 severity and the nasopharyngeal microbiome.
Methods: We conducted a prospective cohort study in Mexico City, collecting nasopharyngeal swabs from 30 COVID-19 patients and 14 healthy volunteers. Microbiome profiling was performed using 16S rRNA gene analysis. Taxonomic assignment, classification, diversity analysis, core microbiome analysis, and statistical analysis were conducted using R packages.
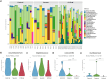
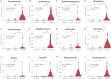
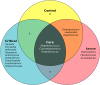
Results: The microbiome data analysis revealed taxonomic shifts within the nasopharyngeal microbiome in severe COVID-19. Particularly, we observed a significant reduction in the relative abundance of Lawsonella and Cutibacterium genera in critically ill COVID-19 patients (p < 0.001). In contrast, these patients exhibited a marked enrichment of Streptococcus, Actinomyces, Peptostreptococcus, Atopobium, Granulicatella, Mogibacterium, Veillonella, Prevotella_7, Rothia, Gemella, Alloprevotella, and Solobacterium genera (p < 0.01). Analysis of the core microbiome across all samples consistently identified the presence of Staphylococcus, Corynebacterium, and Streptococcus.
Conclusions: Our study suggests that the disruption of physicochemical conditions and barriers resulting from inflammatory processes and the intubation procedure in critically ill COVID-19 patients may facilitate the colonization and invasion of the nasopharynx by oral microorganisms.
Keywords: COVID-19; Corynebacterium; Cutibacterium Core microbiome; Intubation; Lawsonella; Nasopharyngeal microbiome; Nasopharynx; Staphylococcus; Streptococcus.
© 2024 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

Similar articles
-
Comparison of the respiratory tract microbiome in hospitalized COVID-19 patients with different disease severity.J Med Virol. 2022 Nov;94(11):5284-5293. doi: 10.1002/jmv.28002. Epub 2022 Jul 26. J Med Virol. 2022. PMID: 35838111 Free PMC article.
-
Metagenomic Next-Generation Sequencing of Nasopharyngeal Specimens Collected from Confirmed and Suspect COVID-19 Patients.mBio. 2020 Nov 20;11(6):e01969-20. doi: 10.1128/mBio.01969-20. mBio. 2020. PMID: 33219095 Free PMC article.
-
Analysis of 16S rRNA Gene Sequence of Nasopharyngeal Exudate Reveals Changes in Key Microbial Communities Associated with Aging.Int J Mol Sci. 2023 Feb 18;24(4):4127. doi: 10.3390/ijms24044127. Int J Mol Sci. 2023. PMID: 36835535 Free PMC article.
-
The salivary and nasopharyngeal microbiomes are associated with SARS-CoV-2 infection and disease severity.J Med Virol. 2023 Feb;95(2):e28445. doi: 10.1002/jmv.28445. J Med Virol. 2023. PMID: 36583481 Free PMC article.
-
The alterations of oral, airway and intestine microbiota in chronic obstructive pulmonary disease: a systematic review and meta-analysis.Front Immunol. 2024 May 8;15:1407439. doi: 10.3389/fimmu.2024.1407439. eCollection 2024. Front Immunol. 2024. PMID: 38779669 Free PMC article.
References
-
- Ren L., Wang Y., Zhong J., Li X., Xiao Y., Li J., Yang J., Fan G., Guo L., Shen Z., Kang L., Shi L., Li Q., Li J., Di L., Li H., Wang C., Wang Y., Wang X., Zou X., Rao J., Zhang L., Wang J., Huang Y., Cao B., Wang J., Li M. Dynamics of the upper respiratory tract microbiota and its association with mortality in COVID-19. Am. J. Respir. Crit. Care Med. 2021;204:1379–1390. doi: 10.1164/rccm.202103-0814OC. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

