Cellular memory function from 3D to 2D: Three-dimensional high density collagen microfiber cultures induce their resistance to reactive oxygen species
- PMID: 38827038
- PMCID: PMC11140783
- DOI: 10.1016/j.mtbio.2024.101097
Cellular memory function from 3D to 2D: Three-dimensional high density collagen microfiber cultures induce their resistance to reactive oxygen species
Abstract
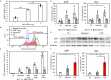
Cell properties generally change when the culture condition is changed. However, mesenchymal stem cells cultured on a hard material surface maintain their differentiation characteristics even after being cultured on a soft material surface. This phenomenon suggests the possibility of a cell culture material to memorize stem cell function even in changing cell culture conditions. However, there are no reports about cell memory function in three-dimensional (3D) culture. In this study, colon cancer cells were cultured with collagen microfibers (CMF) in 3D to evaluate their resistance to reactive oxygen species (ROS) in comparison with a monolayer (2D) culture condition and to understand the effect of 3D-culture on cell memory function. The ratio of ROS-negative cancer cells in 3D culture increased with increasing amounts of CMF and the highest amount of CMF was revealed to be 35-fold higher than that of the 2D condition. The ROS-negative cells ratio was maintained for 7 days after re-seeding in a 2D culture condition, suggesting a 3D-memory function of ROS resistance. The findings of this study will open up new opportunities for 3D culture to induce cell memory function.
Keywords: 3D culture; Cancer tissue; Cellular memory function; Collagen microfibers; Reactive oxygen species.
© 2024 The Authors.
Conflict of interest statement
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Michiya Matsusaki reports financial support was provided by JST-Mirai Program. Michiya MATSUSAKI reports a relationship with JST-Mirai Program that includes: funding grants. Michiya MATSUSAKI reports a relationship with COI-NEXT that includes: funding grants. Michiya MATSUSAKI reports a relationship with New Energy and Industrial Technology Development Organization that includes: funding grants. Michiya MATSUSAKI reports a relationship with Grant-in-Aid for Scientific Research (A) that includes: funding grants. Michiya MATSUSAKI has patent #PCT/JP2022/026062 issued to Assignee. If there are other authors, they declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures






References
-
- Silver J.S., Günay K.A., Cutler A.A., Vogler T.O., Brown T.E., Pawlikowski B.T., Bednarski O.J., Bannister K.L., Rogowski C.J., Mckay A.G., DelRio F.W., Olwin B.B., Anseth K.S. Injury-mediated stiffening persistently activates muscle stem cells through YAP and TAZ mechanotransduction. Sci. Adv. 2021;7 doi: 10.1126/sciadv.abe4501. - DOI - PMC - PubMed
-
- Walker C.J., Crocini C., Ramirez D., Killaars A.R., Grim J.C., Aguado B.A., Clark K., Allen M.A., Dowell R.D., Leinwand L.A., Anseth K.S. Nuclear mechanosensing drives chromatin remodelling in persistently activated fibroblasts. Nat. Biomed. Eng. 2021;5:1485–1499. doi: 10.1038/s41551-021-00709-w. - DOI - PMC - PubMed
-
- Kawano S., Kojima M., Higuchi Y., Sugimoto M., Ikeda K., Sakuyama N., Takahashi S., Hayashi R., Ochiai A., Saito N. Assessment of elasticity of colorectal cancer tissue, clinical utility, pathological and phenotypical relevance. Cancer Sci. 2015;106:1232–1239. doi: 10.1111/cas.12720. - DOI - PMC - PubMed
-
- Shen Y., Wang X., Lu J., Salfenmoser M., Wirsik N.M., Schleussner N., Imle A., Freire Valls A., Radhakrishnan P., Liang J., Wang G., Muley T., Schneider M., Ruiz De Almodovar C., Diz-Muñoz A., Schmidt T. Reduction of liver metastasis stiffness improves response to bevacizumab in metastatic colorectal cancer. Cancer Cell. 2020;37:800–817.e7. doi: 10.1016/j.ccell.2020.05.005. - DOI - PubMed
LinkOut - more resources
Full Text Sources

