PHLDA2 reshapes the immune microenvironment and induces drug resistance in hepatocellular carcinoma
- PMID: 38827322
- PMCID: PMC11136693
- DOI: 10.32604/or.2024.047078
PHLDA2 reshapes the immune microenvironment and induces drug resistance in hepatocellular carcinoma
Abstract
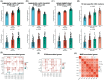
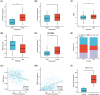
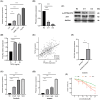
Hepatocellular carcinoma (HCC) is a malignancy known for its unfavorable prognosis. The dysregulation of the tumor microenvironment (TME) can affect the sensitivity to immunotherapy or chemotherapy, leading to treatment failure. The elucidation of PHLDA2's involvement in HCC is imperative, and the clinical value of PHLDA2 is also underestimated. Here, bioinformatics analysis was performed in multiple cohorts to explore the phenotype and mechanism through which PHLDA2 may affect the progression of HCC. Then, the expression and function of PHLDA2 were examined via the qRT-PCR, Western Blot, and MTT assays. Our findings indicate a substantial upregulation of PHLDA2 in HCC, correlated with a poorer prognosis. The methylation levels of PHLDA2 were found to be lower in HCC tissues compared to normal liver tissues. Besides, noteworthy associations were observed between PHLDA2 expression and immune infiltration in HCC. In addition, PHLDA2 upregulation is closely associated with stemness features and immunotherapy or chemotherapy resistance in HCC. In vitro experiments showed that sorafenib or cisplatin significantly up-regulated PHLDA2 mRNA levels, and PHLDA2 knockdown markedly decreased the sensitivity of HCC cells to chemotherapy drugs. Meanwhile, we found that TGF-β induced the expression of PHLDA2 in vitro. The GSEA and in vitro experiment indicated that PHLDA2 may promote the HCC progression via activating the AKT signaling pathway. Our study revealed the novel role of PHLDA2 as an independent prognostic factor, which plays an essential role in TME remodeling and treatment resistance in HCC.
Keywords: Drug resistance; HCC; Immune infiltration; PHLDA2; TME.
© 2024 Feng et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest to report regarding the present study.
Figures




Similar articles
-
A novel mitochondrial unfolded protein response-related risk signature to predict prognosis, immunotherapy and sorafenib sensitivity in hepatocellular carcinoma.Apoptosis. 2024 Jun;29(5-6):768-784. doi: 10.1007/s10495-024-01945-6. Epub 2024 Mar 17. Apoptosis. 2024. PMID: 38493408
-
ALG3 as a prognostic biomarker and mediator of PD-1 blockade resistance in hepatocellular carcinoma.Front Immunol. 2025 May 22;16:1589153. doi: 10.3389/fimmu.2025.1589153. eCollection 2025. Front Immunol. 2025. PMID: 40475760 Free PMC article.
-
Upregulation of lncRNA NIFK-AS1 in hepatocellular carcinoma by m6A methylation promotes disease progression and sorafenib resistance.Hum Cell. 2021 Nov;34(6):1800-1811. doi: 10.1007/s13577-021-00587-z. Epub 2021 Aug 10. Hum Cell. 2021. PMID: 34374933 Review.
-
LncRNA TTN-AS1 intensifies sorafenib resistance in hepatocellular carcinoma by sponging miR-16-5p and upregulation of cyclin E1.Biomed Pharmacother. 2021 Jan;133:111030. doi: 10.1016/j.biopha.2020.111030. Epub 2020 Nov 28. Biomed Pharmacother. 2021. PMID: 33378944
-
Targeting TIME in advanced hepatocellular carcinoma: Mechanisms of drug resistance and treatment strategies.Crit Rev Oncol Hematol. 2025 Jul;211:104735. doi: 10.1016/j.critrevonc.2025.104735. Epub 2025 Apr 16. Crit Rev Oncol Hematol. 2025. PMID: 40250780 Review.
References
-
- Kim, H. I., An, J., Kim, J. Y., Shin, H. P., Park, S. Y.et al. (2022). Postresection period-specific hazard of recurrence as a framework for surveillance strategy in patients with hepatocellular carcinoma: A multicenter outcome study. Liver Cancer , 11(2), 141–151. 10.1159/000518837; - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
