Comparative genome analysis reveals driving forces behind Monkeypox virus evolution and sheds light on the role of ATC trinucleotide motif
- PMID: 38827420
- PMCID: PMC11141602
- DOI: 10.1093/ve/veae043
Comparative genome analysis reveals driving forces behind Monkeypox virus evolution and sheds light on the role of ATC trinucleotide motif
Abstract
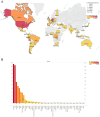
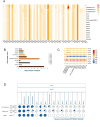
Monkeypox (MPOX), a zoonotic disease originating in Western and Central Africa in 1970, has seen a recent surge in outbreaks across 100+ countries. A comparative analysis of 404 Monkeypox virus (MPXV) genomes revealed notable changes in microsatellite abundance and density, especially within Clades I, IIa, and IIb. Each clade exhibited unique microsatellite motifs, with twenty-six conserved loci specific to MPXV, suggesting their potential as molecular markers in diagnostics. Additionally, nine genes in the MPXV genome featured ten variable hotspot microsatellite regions associated with surface protein synthesis and host control. Notably, gene OPG153, especially at the SSR locus '(ATC)n', exhibited the most pronounced variations among lineages over time and plays a role in virus pathogenesis within the host cell. These findings not only enhance our understanding of MPXV unique molecular profile but also offer valuable insights into potential pathogenic and evolutionary implications.
Keywords: human pathogen; microsatellite; monkeypox; motifs; repeats.
© The Author(s) 2024. Published by Oxford University Press.
Conflict of interest statement
The authors wish to state that they possess no conflicts of interest pertaining to this study, and there exists no substantial financial backing that might have impacted the results of this research.
Figures




Similar articles
-
Differences in pathogenicity among the mpox virus clades: impact on drug discovery and vaccine development.Trends Pharmacol Sci. 2023 Oct;44(10):719-739. doi: 10.1016/j.tips.2023.08.003. Epub 2023 Sep 4. Trends Pharmacol Sci. 2023. PMID: 37673695 Review.
-
Monkeypox virus genomic accordion strategies.Nat Commun. 2024 Apr 18;15(1):3059. doi: 10.1038/s41467-024-46949-7. Nat Commun. 2024. PMID: 38637500 Free PMC article.
-
Virological characterization of the 2022 outbreak-causing monkeypox virus using human keratinocytes and colon organoids.J Med Virol. 2023 Jun;95(6):e28827. doi: 10.1002/jmv.28827. J Med Virol. 2023. PMID: 37278443
-
Mapping global zoonotic niche and interregional transmission risk of monkeypox: a retrospective observational study.Global Health. 2023 Aug 17;19(1):58. doi: 10.1186/s12992-023-00959-0. Global Health. 2023. PMID: 37592305 Free PMC article.
-
Concurrent Clade I and Clade II Monkeypox Virus Circulation, Cameroon, 1979-2022.Emerg Infect Dis. 2024 Mar;30(3):432-443. doi: 10.3201/eid3003.230861. Epub 2024 Feb 7. Emerg Infect Dis. 2024. PMID: 38325363 Free PMC article. Review.
Cited by
-
Identification of potential antigenic proteins and epitopes for the development of a monkeypox virus vaccine: an in silico approach.Mol Divers. 2024 Nov 15. doi: 10.1007/s11030-024-11033-1. Online ahead of print. Mol Divers. 2024. PMID: 39546220
-
Mpox: Global epidemic situation and countermeasures.Virulence. 2025 Dec;16(1):2457958. doi: 10.1080/21505594.2025.2457958. Epub 2025 Feb 8. Virulence. 2025. PMID: 39921615 Free PMC article. Review.
-
Artificial Intelligence (AI) tools to investigate virulence and evaluate the infectivity of Mpox.EXCLI J. 2025 Mar 6;24:403-406. doi: 10.17179/excli2025-8129. eCollection 2025. EXCLI J. 2025. PMID: 40166429 Free PMC article. No abstract available.
References
-
- Alotaibi N. M. et al. (2023) ‘Comparative Genomics Reveals the Presence of Simple Sequence Repeats in Genes Related to Virulence in Plant Pathogenic Pythium Ultimum and Pythium Vexans’, Archives of Microbiology, 205: 256. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

