Analysis of gene expression in microglial apoptotic cell clearance following spinal cord injury based on machine learning algorithms
- PMID: 38827468
- PMCID: PMC11140288
- DOI: 10.3892/etm.2024.12581
Analysis of gene expression in microglial apoptotic cell clearance following spinal cord injury based on machine learning algorithms
Abstract
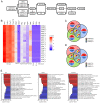
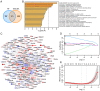
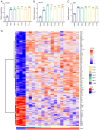
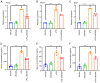
Spinal cord injury (SCI) is a severe neurological complication following spinal fracture, which has long posed a challenge for clinicians. Microglia play a dual role in the pathophysiological process after SCI, both beneficial and detrimental. The underlying mechanisms of microglial actions following SCI require further exploration. The present study combined three different machine learning algorithms, namely weighted gene co-expression network analysis, random forest analysis and least absolute shrinkage and selection operator analysis, to screen for differentially expressed genes in the GSE96055 microglia dataset after SCI. It then used protein-protein interaction networks and gene set enrichment analysis with single genes to investigate the key genes and signaling pathways involved in microglial function following SCI. The results indicated that microglia not only participate in neuroinflammation but also serve a significant role in the clearance mechanism of apoptotic cells following SCI. Notably, bioinformatics analysis and lipopolysaccharide + UNC569 (a MerTK-specific inhibitor) stimulation of BV2 cell experiments showed that the expression levels of Anxa2, Myo1e and Spp1 in microglia were significantly upregulated following SCI, thus potentially involved in regulating the clearance mechanism of apoptotic cells. The present study suggested that Anxa2, Myo1e and Spp1 may serve as potential targets for the future treatment of SCI and provided a theoretical basis for the development of new methods and drugs for treating SCI.
Keywords: apoptotic cells; bioinformatics; least absolute shrinkage and selection operator analysis; machine learning; microglia; protein-protein interaction; random forest analysis; spinal cord injury; weighted gene co-expression network analysis.
Copyright: © 2024 Yan et al.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
Hspb1 and Lgals3 in spinal neurons are closely associated with autophagy following excitotoxicity based on machine learning algorithms.PLoS One. 2024 May 10;19(5):e0303235. doi: 10.1371/journal.pone.0303235. eCollection 2024. PLoS One. 2024. PMID: 38728287 Free PMC article.
-
Identification and validation of biomarkers associated with mitochondrial dysfunction and ferroptosis in rat spinal cord injury.Front Neurol. 2025 Mar 17;16:1526966. doi: 10.3389/fneur.2025.1526966. eCollection 2025. Front Neurol. 2025. PMID: 40166636 Free PMC article.
-
TLR4 aggravates microglial pyroptosis by promoting DDX3X-mediated NLRP3 inflammasome activation via JAK2/STAT1 pathway after spinal cord injury.Clin Transl Med. 2022 Jun;12(6):e894. doi: 10.1002/ctm2.894. Clin Transl Med. 2022. PMID: 35692100 Free PMC article.
-
Taxifolin attenuates neuroinflammation and microglial pyroptosis via the PI3K/Akt signaling pathway after spinal cord injury.Int Immunopharmacol. 2023 Jan;114:109616. doi: 10.1016/j.intimp.2022.109616. Epub 2022 Dec 22. Int Immunopharmacol. 2023. PMID: 36700780
-
Biomaterial-supported MSC transplantation enhances cell-cell communication for spinal cord injury.Stem Cell Res Ther. 2021 Jan 7;12(1):36. doi: 10.1186/s13287-020-02090-y. Stem Cell Res Ther. 2021. PMID: 33413653 Free PMC article. Review.
Cited by
-
Identification of amino acid metabolism‑related genes as diagnostic and prognostic biomarkers in sepsis through machine learning.Exp Ther Med. 2024 Dec 20;29(2):36. doi: 10.3892/etm.2024.12786. eCollection 2025 Feb. Exp Ther Med. 2024. PMID: 39776890 Free PMC article.
References
-
- Thomaz SR, Cipriano G Jr, Formiga MF, Fachin-Martins E, Cipriano GFB, Martins WR, Cahalin LP. Effect of electrical stimulation on muscle atrophy and spasticity in patients with spinal cord injury-a systematic review with meta-analysis. Spinal Cord. 2019;57:258–266. doi: 10.1038/s41393-019-0250-z. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
