Immunoinformatics-Based Design of Multi-epitope DNA and mRNA Vaccines Against Zika Virus
- PMID: 38827811
- PMCID: PMC11143849
- DOI: 10.1177/11779322241257037
Immunoinformatics-Based Design of Multi-epitope DNA and mRNA Vaccines Against Zika Virus
Abstract
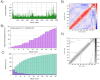
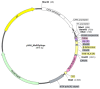
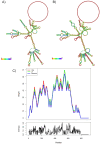
In this study, we used an immunoinformatics approach to predict antigenic epitopes of Zika virus (ZIKV) proteins to assist in designing a vaccine antigen against ZIKV. We performed the prediction of CD8+ T-lymphocyte and antigenic B-cell epitopes of ZIKV proteins. The binding interactions of T-cell epitopes with major histocompatibility complex class I (MHC-I) proteins were assessed. We selected the antigenic, conserved, nontoxic, and immunogenic epitopes, which indicated significant interactions with the human leucocyte antigen (HLA-A and HLA-B) alleles and worldwide population coverage of 76.35%. The predicted epitopes were joined with the help of linkers and an adjuvant. The vaccine antigen was then analyzed through molecular docking with TLR3 and TLR8, and it was in silico cloned in the pVAX1 vector to be used as a DNA vaccine and designed as a mRNA vaccine.
Keywords: Zika virus; immunoinformatics; peptide vaccine.
© The Author(s) 2024.
Conflict of interest statement
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Figures

Similar articles
-
Immunoinformatics Vaccine Design for Zika Virus.Methods Mol Biol. 2023;2673:411-429. doi: 10.1007/978-1-0716-3239-0_28. Methods Mol Biol. 2023. PMID: 37258930
-
A novel vaccine construct against Zika virus fever: insights from epitope-based vaccine discovery through molecular modeling and immunoinformatics approaches.Front Immunol. 2024 Jul 1;15:1426496. doi: 10.3389/fimmu.2024.1426496. eCollection 2024. Front Immunol. 2024. PMID: 39050858 Free PMC article.
-
In silico construction of a multiepitope Zika virus vaccine using immunoinformatics tools.Sci Rep. 2022 Jan 7;12(1):53. doi: 10.1038/s41598-021-03990-6. Sci Rep. 2022. PMID: 34997041 Free PMC article.
-
Immunoinformatics guided rational design of a next generation multi epitope based peptide (MEBP) vaccine by exploring Zika virus proteome.Infect Genet Evol. 2020 Jun;80:104199. doi: 10.1016/j.meegid.2020.104199. Epub 2020 Jan 18. Infect Genet Evol. 2020. PMID: 31962160
-
Epitope Prediction by Novel Immunoinformatics Approach: A State-of-the-art Review.Int J Pept Res Ther. 2020;26(2):1155-1163. doi: 10.1007/s10989-019-09918-z. Epub 2019 Aug 20. Int J Pept Res Ther. 2020. PMID: 32435171 Free PMC article. Review.
Cited by
-
An mRNA vaccine candidate encoding cholera toxin subunit B and conserved antigens of influenza viruses confers cross-protection against influenza a viruses in adult and aged mice.Hum Vaccin Immunother. 2025 Dec;21(1):2453304. doi: 10.1080/21645515.2025.2453304. Epub 2025 Feb 16. Hum Vaccin Immunother. 2025. PMID: 39957235 Free PMC article.
-
Unleashing the future of cancer immunotherapy: in silico design of a multi-epitope and mRNA vaccine duo targeting EWSR1-ATF1, EWSR1-CREB1, and PRAME to conquer clear cell sarcoma using immunoinformatics approaches.Med Oncol. 2025 Jun 28;42(8):295. doi: 10.1007/s12032-025-02863-6. Med Oncol. 2025. PMID: 40581703
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

