Expanding the horizons of targeted protein degradation: A non-small molecule perspective
- PMID: 38828146
- PMCID: PMC11143490
- DOI: 10.1016/j.apsb.2024.01.010
Expanding the horizons of targeted protein degradation: A non-small molecule perspective
Abstract
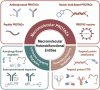
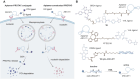
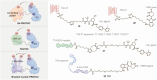
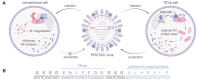
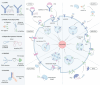
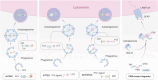
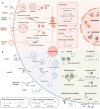
Targeted protein degradation (TPD) represented by proteolysis targeting chimeras (PROTACs) marks a significant stride in drug discovery. A plethora of innovative technologies inspired by PROTAC have not only revolutionized the landscape of TPD but have the potential to unlock functionalities beyond degradation. Non-small-molecule-based approaches play an irreplaceable role in this field. A wide variety of agents spanning a broad chemical spectrum, including peptides, nucleic acids, antibodies, and even vaccines, which not only prove instrumental in overcoming the constraints of conventional small molecule entities but also provided rapidly renewing paradigms. Herein we summarize the burgeoning non-small molecule technological platforms inspired by PROTACs, including three major trajectories, to provide insights for the design strategies based on novel paradigms.
Keywords: Autophagy; Endocytosis; Post-translational modification; Proteolysis targeting chimera; Proximity-inducing modality; Targeted protein degradation.
© 2024 The Authors.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures





Similar articles
-
Recent breakthroughs in innovative elements, multidimensional enhancements, derived technologies, and novel applications of PROTACs.Biomed Pharmacother. 2024 Nov;180:117584. doi: 10.1016/j.biopha.2024.117584. Epub 2024 Oct 19. Biomed Pharmacother. 2024. PMID: 39427546 Review.
-
Advancement of targeted protein degradation strategies as therapeutics for undruggable disease targets.Future Med Chem. 2023 May;15(10):867-883. doi: 10.4155/fmc-2023-0072. Epub 2023 May 31. Future Med Chem. 2023. PMID: 37254917 Review.
-
Novel strategies and promising opportunities for targeted protein degradation: An innovative therapeutic approach to overcome cancer resistance.Pharmacol Ther. 2023 Apr;244:108371. doi: 10.1016/j.pharmthera.2023.108371. Epub 2023 Mar 5. Pharmacol Ther. 2023. PMID: 36871783 Review.
-
Advancing targeted protein degradation for metabolic diseases therapy.Pharmacol Res. 2023 Feb;188:106627. doi: 10.1016/j.phrs.2022.106627. Epub 2022 Dec 21. Pharmacol Res. 2023. PMID: 36566001 Review.
-
Targeted protein degradation in cancers: Orthodox PROTACs and beyond.Innovation (Camb). 2023 Mar 15;4(3):100413. doi: 10.1016/j.xinn.2023.100413. eCollection 2023 May 15. Innovation (Camb). 2023. PMID: 37033156 Free PMC article. Review.
Cited by
-
A novel ROR1-targeting antibody-PROTAC conjugate promotes BRD4 degradation for solid tumor treatment.Theranostics. 2025 Jan 1;15(4):1238-1254. doi: 10.7150/thno.102531. eCollection 2025. Theranostics. 2025. PMID: 39816690 Free PMC article.
-
Interplay of PROTAC Complex Dynamics for Undruggable Targets: Insights into Ternary Complex Behavior and Linker Design.ACS Med Chem Lett. 2024 Jul 29;15(8):1306-1318. doi: 10.1021/acsmedchemlett.4c00189. eCollection 2024 Aug 8. ACS Med Chem Lett. 2024. PMID: 39140051 Free PMC article.
-
Treating ICB-resistant cancer by inhibiting PD-L1 via DHHC3 degradation induced by cell penetrating peptide-induced chimera conjugates.Cell Death Dis. 2024 Sep 30;15(9):701. doi: 10.1038/s41419-024-07073-y. Cell Death Dis. 2024. PMID: 39349454 Free PMC article.
-
Optineurin restrains CCR7 degradation to guide type II collagen-stimulated dendritic cell migration in rheumatoid arthritis.Acta Pharm Sin B. 2025 Mar;15(3):1626-1642. doi: 10.1016/j.apsb.2025.02.004. Epub 2025 Feb 11. Acta Pharm Sin B. 2025. PMID: 40370566 Free PMC article.
-
Development of miRNA-based PROTACs targeting Lin28 for breast cancer therapy.Sci Adv. 2024 Sep 20;10(38):eadp0334. doi: 10.1126/sciadv.adp0334. Epub 2024 Sep 18. Sci Adv. 2024. PMID: 39292784 Free PMC article.
References
Publication types
LinkOut - more resources
Full Text Sources

