Carrimycin inhibits coronavirus replication by decreasing the efficiency of programmed -1 ribosomal frameshifting through directly binding to the RNA pseudoknot of viral frameshift-stimulatory element
- PMID: 38828157
- PMCID: PMC11143517
- DOI: 10.1016/j.apsb.2024.02.023
Carrimycin inhibits coronavirus replication by decreasing the efficiency of programmed -1 ribosomal frameshifting through directly binding to the RNA pseudoknot of viral frameshift-stimulatory element
Abstract
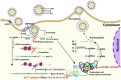
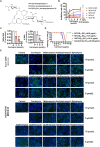
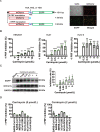
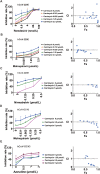
The pandemic of SARS-CoV-2 worldwide with successive emerging variants urgently calls for small-molecule oral drugs with broad-spectrum antiviral activity. Here, we show that carrimycin, a new macrolide antibiotic in the clinic and an antiviral candidate for SARS-CoV-2 in phase III trials, decreases the efficiency of programmed -1 ribosomal frameshifting of coronaviruses and thus impedes viral replication in a broad-spectrum fashion. Carrimycin binds directly to the coronaviral frameshift-stimulatory element (FSE) RNA pseudoknot, interrupting the viral protein translation switch from ORF1a to ORF1b and thereby reducing the level of the core components of the viral replication and transcription complexes. Combined carrimycin with known viral replicase inhibitors yielded a synergistic inhibitory effect on coronaviruses. Because the FSE mechanism is essential in all coronaviruses, carrimycin could be a new broad-spectrum antiviral drug for human coronaviruses by directly targeting the conserved coronaviral FSE RNA. This finding may open a new direction in antiviral drug discovery for coronavirus variants.
Keywords: Antiviral agent; Broad-spectrum antiviral activity; Carrimycin; Coronavirus; Programmed –1 ribosomal frameshifting; RNA pseudoknot; RNA target; Synergistic inhibitory effect.
© 2024 The Authors.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


Similar articles
-
Carrimycin exhibited broad spectrum inhibitory activities against coronaviruses replication through down-regulating host factor TMEM41B.Acta Pharmacol Sin. 2025 Jul;46(7):2006-2015. doi: 10.1038/s41401-025-01577-9. Epub 2025 May 15. Acta Pharmacol Sin. 2025. PMID: 40374896
-
Synthesis and evolution of 16-membered macrolide carrimycin derivatives as a novel class of anti-HCoV-OC43 agents targeting viral FSE RNA.Eur J Med Chem. 2025 Apr 5;287:117373. doi: 10.1016/j.ejmech.2025.117373. Epub 2025 Feb 7. Eur J Med Chem. 2025. PMID: 39952097
-
A Novel Frameshifting Inhibitor Having Antiviral Activity against Zoonotic Coronaviruses.Viruses. 2021 Aug 18;13(8):1639. doi: 10.3390/v13081639. Viruses. 2021. PMID: 34452503 Free PMC article.
-
The frameshifting element in coronaviruses: structure, function, and potential as a therapeutic target.Trends Pharmacol Sci. 2025 Jun;46(6):535-550. doi: 10.1016/j.tips.2025.04.003. Epub 2025 May 16. Trends Pharmacol Sci. 2025. PMID: 40382241 Review.
-
Targeting Ribosomal Frameshifting as an Antiviral Strategy: From HIV-1 to SARS-CoV-2.Acc Chem Res. 2021 Sep 7;54(17):3349-3361. doi: 10.1021/acs.accounts.1c00316. Epub 2021 Aug 17. Acc Chem Res. 2021. PMID: 34403258 Review.
Cited by
-
Mycophenolate mofetil exerts broad-spectrum antiviral activity against coronaviruses including SARS-CoV-2.Virol J. 2025 Mar 4;22(1):56. doi: 10.1186/s12985-025-02673-2. Virol J. 2025. PMID: 40038695 Free PMC article.
-
The synergistic antitumour effect of Carrimycin combined with 5-fluorouracil on colorectal cancer.Sci Rep. 2025 Mar 17;15(1):9155. doi: 10.1038/s41598-025-94306-5. Sci Rep. 2025. PMID: 40097541 Free PMC article.
-
Progress in Research on Inhibitors Targeting SARS-CoV-2 Main Protease (Mpro).ACS Omega. 2024 Aug 2;9(32):34196-34219. doi: 10.1021/acsomega.4c03023. eCollection 2024 Aug 13. ACS Omega. 2024. PMID: 39157135 Free PMC article. Review.
-
Stem loop binding protein promotes SARS-CoV-2 replication via -1 programmed ribosomal frameshifting.Signal Transduct Target Ther. 2025 Jun 13;10(1):192. doi: 10.1038/s41392-025-02277-w. Signal Transduct Target Ther. 2025. PMID: 40514371 Free PMC article.
-
Carrimycin exhibited broad spectrum inhibitory activities against coronaviruses replication through down-regulating host factor TMEM41B.Acta Pharmacol Sin. 2025 Jul;46(7):2006-2015. doi: 10.1038/s41401-025-01577-9. Epub 2025 May 15. Acta Pharmacol Sin. 2025. PMID: 40374896
References
-
- Food and Drug Administration . 2023. Coronavirus (COVID-19)|Drugs.https://www.fda.gov/drugs/emergency-preparedness-drugs/coronavirus-covid... Available from:
LinkOut - more resources
Full Text Sources
Miscellaneous

