Plasmodium vivax spleen-dependent protein 1 and its role in extracellular vesicles-mediated intrasplenic infections
- PMID: 38828264
- PMCID: PMC11140020
- DOI: 10.3389/fcimb.2024.1408451
Plasmodium vivax spleen-dependent protein 1 and its role in extracellular vesicles-mediated intrasplenic infections
Abstract
Recent studies indicate that human spleen contains over 95% of the total parasite biomass during chronic asymptomatic infections caused by Plasmodium vivax. Previous studies have demonstrated that extracellular vesicles (EVs) secreted from infected reticulocytes facilitate binding to human spleen fibroblasts (hSFs) and identified parasite genes whose expression was dependent on an intact spleen. Here, we characterize the P. vivax spleen-dependent hypothetical gene (PVX_114580). Using CRISPR/Cas9, PVX_114580 was integrated into P. falciparum 3D7 genome and expressed during asexual stages. Immunofluorescence analysis demonstrated that the protein, which we named P. vivax Spleen-Dependent Protein 1 (PvSDP1), was located at the surface of infected red blood cells in the transgenic line and this localization was later confirmed in natural infections. Plasma-derived EVs from P. vivax-infected individuals (PvEVs) significantly increased cytoadherence of 3D7_PvSDP1 transgenic line to hSFs and this binding was inhibited by anti-PvSDP1 antibodies. Single-cell RNAseq of PvEVs-treated hSFs revealed increased expression of adhesion-related genes. These findings demonstrate the importance of parasite spleen-dependent genes and EVs from natural infections in the formation of intrasplenic niches in P. vivax, a major challenge for malaria elimination.
Keywords: CRISPR/Ca9; Plasmodium vivax; extracellular vesicles (EVs); intrasplenic infections; single-cell RNASeq (scRNASeq); spleen fibroblasts.
Copyright © 2024 Ayllon-Hermida, Nicolau-Fernandez, Larrinaga, Aparici-Herraiz, Tintó-Font, Llorà-Batlle, Orban, Yasnot, Graupera, Esteller, Popovici, Cortés, del Portillo and Fernandez-Becerra.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
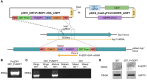
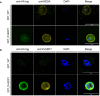
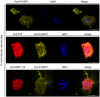
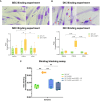
References
-
- Aparici-Herraiz I., Gualdrón-López M., Castro-Cavadía C. J., Carmona-Fonseca J., Yasnot M. F., Fernandez-Becerra C., et al. (2022). Antigen discovery in circulating extracellular vesicles from plasmodium vivax patients. Front. Cell Infect. Microbiol. 11. doi: 10.3389/fcimb.2021.811390 - DOI - PMC - PubMed
-
- Bernabeu M., Lopez F. J., Ferrer M., Martin-Jaular L., Razaname A., Corradin G., et al. (2012). Functional analysis of Plasmodium vivax VIR proteins reveals different subcellular localizations and cytoadherence to the ICAM-1 endothelial receptor. Cell Microbiol. 14, 386–400. doi: 10.1111/j.1462-5822.2011.01726.x - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

