Non-canonical mRNA translation initiation in cell stress and cancer
- PMID: 38828390
- PMCID: PMC11140632
- DOI: 10.1093/narcan/zcae026
Non-canonical mRNA translation initiation in cell stress and cancer
Abstract
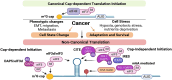
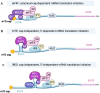
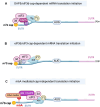
The now well described canonical mRNA translation initiation mechanism of m7G 'cap' recognition by cap-binding protein eIF4E and assembly of the canonical pre-initiation complex consisting of scaffolding protein eIF4G and RNA helicase eIF4A has historically been thought to describe all cellular mRNA translation. However, the past decade has seen the discovery of alternative mechanisms to canonical eIF4E mediated mRNA translation initiation. Studies have shown that non-canonical alternate mechanisms of cellular mRNA translation initiation, whether cap-dependent or independent, serve to provide selective translation of mRNAs under cell physiological and pathological stress conditions. These conditions typically involve the global downregulation of canonical eIF4E1/cap-mediated mRNA translation, and selective translational reprogramming of the cell proteome, as occurs in tumor development and malignant progression. Cancer cells must be able to maintain physiological plasticity to acquire a migratory phenotype, invade tissues, metastasize, survive and adapt to severe microenvironmental stress conditions that involve inhibition of canonical mRNA translation initiation. In this review we describe the emerging, important role of non-canonical, alternate mechanisms of mRNA translation initiation in cancer, particularly in adaptation to stresses and the phenotypic cell fate changes involved in malignant progression and metastasis. These alternate translation initiation mechanisms provide new targets for oncology therapeutics development.
© The Author(s) 2024. Published by Oxford University Press on behalf of NAR Cancer.
Figures

References
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

