Genetic variation and evolutionary characteristics of Echovirus 11: new variant within genotype D5 associated with neonatal death found in China
- PMID: 38828746
- PMCID: PMC11159588
- DOI: 10.1080/22221751.2024.2361814
Genetic variation and evolutionary characteristics of Echovirus 11: new variant within genotype D5 associated with neonatal death found in China
Abstract
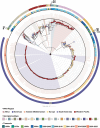
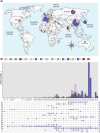
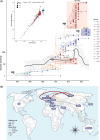
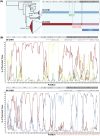
Echovirus 11 (E11) has gained attention owing to its association with severe neonatal infections. From 2018 to 2023, a surge in severe neonatal cases and fatalities linked to a novel variant of genotype D5 was documented in China, France, and Italy. However, the prevention and control of E11 variants have been hampered by limited background data on the virus circulation and genetic variance. Therefore, the present study investigated the circulating dynamics of E11 and the genetic variation and molecular evolution of genotype D5 through the collection of strains from the national acute flaccid paralysis (AFP) and hand, foot, and mouth disease (HFMD) surveillance system in China during 2000-2022 and genetic sequences published in the GenBank database. The results of this study revealed a prevalent dynamic of E11 circulation, with D5 being the predominant genotype worldwide. Further phylogenetic analysis of genotype D5 indicated that it could be subdivided into three important geographic clusters (D5-CHN1: 2014-2019, D5-CHN2: 2016-2022, and D5-EUR: 2022-2023). Additionally, variant-specific (144) amino acid mutation sites and positive-selection pressure sites (132, 262) were identified in the VP1 region. Cluster-specific recombination patterns were also identified, with CVB5, E6, and CVB4 as the major recombinant viruses. These findings provide a preliminary landscape of E11 circulation worldwide and basic scientific data for further study of the pathogenicity of E11 variants.
Keywords: Echovirus 11; Enterovirus; gene recombination; molecular epidemiology; variants; virus evolution; virus transmission.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures

Similar articles
-
Molecular characterization of emerging Echovirus 11 (E11) shed light on the recombinant origin of a variant associated with severe hepatitis in neonates.J Med Virol. 2024 May;96(5):e29658. doi: 10.1002/jmv.29658. J Med Virol. 2024. PMID: 38727043
-
Intensified Circulation of Echovirus 11 after the COVID-19 Pandemic in Poland: Detection of a Highly Pathogenic Virus Variant.Viruses. 2024 Jun 24;16(7):1011. doi: 10.3390/v16071011. Viruses. 2024. PMID: 39066174 Free PMC article.
-
Multiple genotypes of Echovirus 11 circulated in mainland China between 1994 and 2017.Sci Rep. 2019 Jul 22;9(1):10583. doi: 10.1038/s41598-019-46870-w. Sci Rep. 2019. PMID: 31332200 Free PMC article.
-
Echovirus 11 lineage I and other enteroviruses in hospitalized children with acute respiratory infection in Southern Italy, 2022- 2023.Int J Infect Dis. 2024 Sep;146:107091. doi: 10.1016/j.ijid.2024.107091. Epub 2024 May 8. Int J Infect Dis. 2024. PMID: 38729515
-
Two dramatically different clinical scenarios of neonatal Echovirus-11 infection in late preterm male twins: a case report and review of the literature.Ital J Pediatr. 2025 Feb 20;51(1):49. doi: 10.1186/s13052-025-01880-5. Ital J Pediatr. 2025. PMID: 39980007 Free PMC article. Review.
Cited by
-
Neonatal acute liver failure cases with echovirus 11 infections, Japan, August to November 2024.Euro Surveill. 2025 Jan;30(1):2400822. doi: 10.2807/1560-7917.ES.2025.30.1.2400822. Euro Surveill. 2025. PMID: 39790073 Free PMC article.
-
Epidemiological, etiological, and serological characteristics of hand, foot, and mouth disease in Guizhou Province, Southwest China, from 2008 to 2023.PLoS Negl Trop Dis. 2025 Aug 18;19(8):e0013394. doi: 10.1371/journal.pntd.0013394. eCollection 2025 Aug. PLoS Negl Trop Dis. 2025. PMID: 40825065 Free PMC article.
References
-
- Khetsuriani N, Lamonte-Fowlkes A, Oberst S, et al. . Enterovirus surveillance–United States, 1970-2005. MMWR Surveill Summ. 2006;55:1–20. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
