Site-specific regulation of RNA editing with ribose-modified nucleoside analogs in ADAR guide strands
- PMID: 38828787
- PMCID: PMC11229365
- DOI: 10.1093/nar/gkae461
Site-specific regulation of RNA editing with ribose-modified nucleoside analogs in ADAR guide strands
Abstract
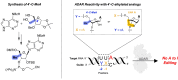
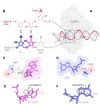
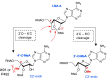
Adenosine Deaminases Acting on RNA (ADARs) are enzymes that catalyze the conversion of adenosine to inosine in RNA duplexes. These enzymes can be harnessed to correct disease-causing G-to-A mutations in the transcriptome because inosine is translated as guanosine. Guide RNAs (gRNAs) can be used to direct the ADAR reaction to specific sites. Chemical modification of ADAR guide strands is required to facilitate delivery, increase metabolic stability, and increase the efficiency and selectivity of the editing reaction. Here, we show the ADAR reaction is highly sensitive to ribose modifications (e.g. 4'-C-methylation and Locked Nucleic Acid (LNA) substitution) at specific positions within the guide strand. Our studies were enabled by the synthesis of RNA containing a new, ribose-modified nucleoside analog (4'-C-methyladenosine). Importantly, the ADAR reaction is potently inhibited by LNA or 4'-C-methylation at different positions in the ADAR guide. While LNA at guide strand positions -1 and -2 block the ADAR reaction, 4'-C-methylation only inhibits at the -2 position. These effects are rationalized using high-resolution structures of ADAR-RNA complexes. This work sheds additional light on the mechanism of ADAR deamination and aids in the design of highly selective ADAR guide strands for therapeutic editing using chemically modified RNA.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures







References
-
- Pullirsch D., Jantsch M.F.. Proteome diversification by adenosine to inosine RNA-editing. RNA Biol. 2010; 7:205–212. - PubMed
-
- Higuchi M., Maas S., Single F.N., Hartner J., Rozov A., Burnashev N., Feldmeyer D., Sprengel R., Seeburg P.H.. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature. 2000; 406:78–81. - PubMed

