TBX3 is essential for establishment of the posterior boundary of anterior genes and upregulation of posterior genes together with HAND2 during the onset of limb bud development
- PMID: 38828908
- PMCID: PMC11190573
- DOI: 10.1242/dev.202722
TBX3 is essential for establishment of the posterior boundary of anterior genes and upregulation of posterior genes together with HAND2 during the onset of limb bud development
Abstract
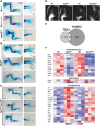
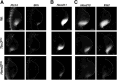
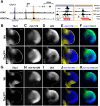
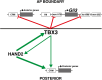
During limb bud formation, axis polarities are established as evidenced by the spatially restricted expression of key regulator genes. In particular, the mutually antagonistic interaction between the GLI3 repressor and HAND2 results in distinct and non-overlapping anterior-distal Gli3 and posterior Hand2 expression domains. This is a hallmark of the establishment of antero-posterior limb axis polarity, together with spatially restricted expression of homeodomain and other transcriptional regulators. Here, we show that TBX3 is required for establishment of the posterior expression boundary of anterior genes in mouse limb buds. ChIP-seq and differential gene expression analysis of wild-type and mutant limb buds identifies TBX3-specific and shared TBX3-HAND2 target genes. High sensitivity fluorescent whole-mount in situ hybridisation shows that the posterior expression boundaries of anterior genes are positioned by TBX3-mediated repression, which excludes anterior genes such as Gli3, Alx4, Hand1 and Irx3/5 from the posterior limb bud mesenchyme. This exclusion delineates the posterior mesenchymal territory competent to establish the Shh-expressing limb bud organiser. In turn, HAND2 is required for Shh activation and cooperates with TBX3 to upregulate shared posterior identity target genes in early limb buds.
Keywords: Gli3; Enhancer activities; Gene expression boundary; Irx genes; Limb bud; Mouse; TBX3 target genes.
© 2024. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures




Similar articles
-
Distinct roles of Hand2 in initiating polarity and posterior Shh expression during the onset of mouse limb bud development.PLoS Genet. 2010 Apr 8;6(4):e1000901. doi: 10.1371/journal.pgen.1000901. PLoS Genet. 2010. PMID: 20386744 Free PMC article.
-
HAND2 targets define a network of transcriptional regulators that compartmentalize the early limb bud mesenchyme.Dev Cell. 2014 Nov 10;31(3):345-357. doi: 10.1016/j.devcel.2014.09.018. Epub 2014 Nov 10. Dev Cell. 2014. PMID: 25453830 Free PMC article.
-
The microRNA-processing enzyme Dicer is dispensable for somite segmentation but essential for limb bud positioning.Dev Biol. 2011 Mar 15;351(2):254-65. doi: 10.1016/j.ydbio.2011.01.005. Epub 2011 Jan 20. Dev Biol. 2011. PMID: 21256124 Free PMC article.
-
How the embryo makes a limb: determination, polarity and identity.J Anat. 2015 Oct;227(4):418-30. doi: 10.1111/joa.12361. Epub 2015 Aug 7. J Anat. 2015. PMID: 26249743 Free PMC article. Review.
-
Dynamic and self-regulatory interactions among gene regulatory networks control vertebrate limb bud morphogenesis.Curr Top Dev Biol. 2020;139:61-88. doi: 10.1016/bs.ctdb.2020.02.005. Epub 2020 Mar 9. Curr Top Dev Biol. 2020. PMID: 32450969 Review.
Cited by
-
Molecular basis of positional memory in limb regeneration.Nature. 2025 Jun;642(8068):730-738. doi: 10.1038/s41586-025-09036-5. Epub 2025 May 21. Nature. 2025. PMID: 40399677 Free PMC article.
-
Comparative single-cell analyses reveal evolutionary repurposing of a conserved gene programme in bat wing development.Nat Ecol Evol. 2025 Jul 16. doi: 10.1038/s41559-025-02780-x. Online ahead of print. Nat Ecol Evol. 2025. PMID: 40670657
References
-
- Aydoğdu, N., Rudat, C., Trowe, M.-O., Kaiser, M., Ludtke, T. H., Taketo, M. M., Christoffels, V. M., Moon, A. and Kispert, A. (2018). TBX2 and TBX3 act downstream of canonical WNT signaling in patterning and differentiation of the mouse ureteric mesenchyme. Development 145, dev.171827. 10.1242/dev.171827 - DOI - PubMed
-
- Bamshad, M., Le, T., Watkins, W. S., Dixon, M. E., Kramer, B. E., Roeder, A. D., Carey, J. C., Root, S., Schinzel, A., Van Maldergem, L.et al. (1999). The spectrum of mutations in TBX3: genotype/phenotype relationship in ulnar-mammary syndrome. Am J. Hum. Genet. 64, 1550-1562. 10.1086/302417 - DOI - PMC - PubMed
-
- Bastida, M. F., Pérez-Gómez, R., Trofka, A., Zhu, J., Rada-Iglesias, A., Sheth, R., Stadler, H. S., Mackem, S. and Ros, M. A. (2020). The formation of the thumb requires direct modulation of gli3 transcription by HOXA13. Proc. Natl. Acad. Sci. USA 117, 1090-1096. 10.1073/pnas.1919470117 - DOI - PMC - PubMed
-
- Benjamini, Y. and Hochberg, Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Method 57, 289-300. 10.1111/j.2517-6161.1995.tb02031.x - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

