Quantification of human papillomavirus cell-free DNA from low-volume blood plasma samples by digital PCR
- PMID: 38829114
- PMCID: PMC11218464
- DOI: 10.1128/spectrum.00024-24
Quantification of human papillomavirus cell-free DNA from low-volume blood plasma samples by digital PCR
Abstract
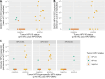
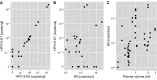
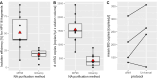
The incidence rate of human papillomavirus-driven oropharyngeal cancer (HPV-OPC) is increasing in countries with high human development index. HPV cell-free DNA (cfDNA) isolated from 3 to 4 mL blood plasma has been successfully used for therapy surveillance. A highly discussed application of HPV-cfDNA is early detection of HPV-OPC. This requires sensitive and specific cfDNA detection as cfDNA levels can be very low. To study the predictive power of pre-diagnostic HPV-cfDNA, archived samples from epidemiological cohorts with limited plasma volume are an important source. To establish a cfDNA detection workflow for low plasma volumes, we compared cfDNA purification methods [MagNA Pure 96 (MP96) and QIAamp ccfDNA/RNA] and digital PCR systems (Biorad QX200 and QIAGEN QIAcuity One). Final assay validation included 65 low-volume plasma samples from oropharyngeal cancer (OPC) patients with defined HPV status stored for 2-9 years. MP96 yielded a 28% higher cfDNA isolation efficiency in comparison to QIAamp. Both digital PCR systems showed comparable analytical sensitivity (6-17 copies for HPV16 and HPV33), but QIAcuity detected both types in the same assay. In the validation set, the assay had 80% sensitivity (n = 28/35) for HPV16 and HPV33 and a specificity of 97% (n = 29/30). In samples with ≥750 µL plasma, the sensitivity was 85% (n = 17/20), while in samples with <750 µL plasma, it was 73% (n = 11/15). Despite the expected drop in sensitivity with decreased plasma volume, the assay is sensitive and highly specific even in low-volume samples and thus suited for studies exploring HPV-cfDNA as an early HPV-OPC detection marker in low-volume archival material.IMPORTANCEHPV-OPC has a favorable prognosis compared to HPV-negative OPC. However, the majority of tumors is diagnosed after regional spread, thus making intensive treatment necessary. This can cause lasting morbidity with a large impact on quality of life. One potential method to decrease treatment-related morbidity is early detection of the cancer. HPV cfDNA has been successfully used for therapy surveillance and has also been detected in pre-diagnostic samples of HPV-OPC patients. These pre-diagnostic samples are only commonly available from biobanks, which usually only have small volumes of blood plasma available. Hence, we have developed a workflow optimized for small-volume archival samples. With this method, a high sensitivity can be achieved despite sample limitations, making it suitable to conduct further large-scale biobank studies of HPV-cfDNA as a prognostic biomarker for HPV-OPC.
Keywords: HPV; OPC; cfDNA; digital PCR; early detection; liquid biopsy.
Conflict of interest statement
T.W. serves on advisory boards for Merck, Sharp & Dohme (MSD). S.L. serves on advisory Boards of MSD, Bristol Myers, Squibb (BMS), and Astra Zeneca (AZ) and has received honoraria from MSD, BMS, AZ, and Merck Serono. D.L.F. has received research funding or in-kind funding from BMS, Calico, Predicine, BostonGene, and NeoGenomics and has received consulting fees from Merck, Noetic, Chrysalis Biomedical Advisors, Arcadia, and Focus. He receives salary support from National Institutes of Health (NIH)/National Institute of Dental and Craniofacial Research (NIDCR) K23 DE029811, NIH/NIDCR R03DE030550, and NIH/National Cancer Institute R21CA267152.
Figures


References
-
- Hong AM, Martin A, Armstrong BK, Lee CS, Jones D, Chatfield MD, Zhang M, Harnett G, Clark J, Elliott M, Milross C, Smee R, Corry J, Liu C, Porceddu S, Vaska K, Veness M, Morgan G, Fogarty G, Veivers D, Rees G, Rose B. 2013. Human papillomavirus modifies the prognostic significance of T stage and possibly N stage in tonsillar cancer. Ann Oncol 24:215–219. doi:10.1093/annonc/mds205 - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

