Intestinal cDC1s provide cues required for CD4+ T cell-mediated resistance to Cryptosporidium
- PMID: 38829369
- PMCID: PMC11148471
- DOI: 10.1084/jem.20232067
Intestinal cDC1s provide cues required for CD4+ T cell-mediated resistance to Cryptosporidium
Abstract
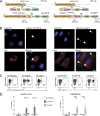
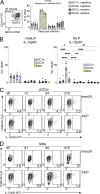
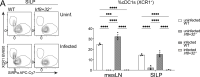
Cryptosporidium is an enteric pathogen and a prominent cause of diarrheal disease worldwide. Control of Cryptosporidium requires CD4+ T cells, but how protective CD4+ T cell responses are generated is poorly understood. Here, Cryptosporidium parasites that express MHCII-restricted model antigens were generated to understand the basis for CD4+ T cell priming and effector function. These studies revealed that parasite-specific CD4+ T cells are primed in the draining mesenteric lymph node but differentiate into Th1 cells in the gut to provide local parasite control. Although type 1 conventional dendritic cells (cDC1s) were dispensable for CD4+ T cell priming, they were required for CD4+ T cell gut homing and were a source of IL-12 at the site of infection that promoted local production of IFN-γ. Thus, cDC1s have distinct roles in shaping CD4+ T cell responses to an enteric infection: first, to promote gut homing from the mesLN, and second, to drive effector responses in the intestine.
© 2024 Cohn et al.
Conflict of interest statement
Disclosures: J.A. Gullicksrud is currently affiliated with Cell Press, but all experiments performed by her for these studies were done before she worked there. Therefore, the authors declare no competing interests.
Figures









Update of
-
Intestinal cDC1s provide IL-12 dependent and independent functions required for CD4+ T cell-mediated resistance to Cryptosporidium.bioRxiv [Preprint]. 2023 Nov 13:2023.11.11.566669. doi: 10.1101/2023.11.11.566669. bioRxiv. 2023. Update in: J Exp Med. 2024 Jul 1;221(7):e20232067. doi: 10.1084/jem.20232067. PMID: 38014026 Free PMC article. Updated. Preprint.
References
-
- Bilate, A.M., London M., Castro T.B.R., Mesin L., Bortolatto J., Kongthong S., Harnagel A., Victora G.D., and Mucida D.. 2020. T cell receptor is required for differentiation, but not maintenance, of intestinal CD4+ intraepithelial lymphocytes. Immunity. 53:1001–1014.e20. 10.1016/j.immuni.2020.09.003 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- T32 AI055428/AI/NIAID NIH HHS/United States
- MFE-176621/CAPMC/ CIHR/Canada
- T32 AI007532/AI/NIAID NIH HHS/United States
- U01AI163671/NH/NIH HHS/United States
- Commonwealth of Pennsylvania
- U01 AI160664/AI/NIAID NIH HHS/United States
- R01 AI157247/AI/NIAID NIH HHS/United States
- P2BEP3_191774/SNSF_/Swiss National Science Foundation/Switzerland
- T32 AI055400/AI/NIAID NIH HHS/United States
- 300355/Fonds de Recherche du Québec-Santé
- T32 AI007632/AI/NIAID NIH HHS/United States
- R01 AI112427/AI/NIAID NIH HHS/United States
- R01 AI148249/AI/NIAID NIH HHS/United States
- U01 AI163671/AI/NIAID NIH HHS/United States
- F30 AI169744/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Research Materials

