Metagenomic exploration of cold-active enzymes for detergent applications: Characterization of a novel, cold-active and alkali-stable GH8 endoglucanase from ikaite columns in SW Greenland
- PMID: 38829370
- PMCID: PMC11146146
- DOI: 10.1111/1751-7915.14466
Metagenomic exploration of cold-active enzymes for detergent applications: Characterization of a novel, cold-active and alkali-stable GH8 endoglucanase from ikaite columns in SW Greenland
Abstract
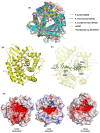
Microbial communities from extreme environments are largely understudied, but are essential as producers of metabolites, including enzymes, for industrial processes. As cultivation of most microorganisms remains a challenge, culture-independent approaches for enzyme discovery in the form of metagenomics to analyse the genetic potential of a community are rapidly becoming the way forward. This study focused on analysing a metagenome from the cold and alkaline ikaite columns in Greenland, identifying 282 open reading frames (ORFs) that encoded putative carbohydrate-modifying enzymes with potential applications in, for example detergents and other processes where activity at low temperature and high pH is desired. Seventeen selected ORFs, representing eight enzyme families were synthesized and expressed in two host organisms, Escherichia coli and Aliivibrio wodanis. Aliivibrio wodanis demonstrated expression of a more diverse range of enzyme classes compared to E. coli, emphasizing the importance of alternative expression systems for enzymes from extremophilic microorganisms. To demonstrate the validity of the screening strategy, we chose a recombinantly expressed cellulolytic enzyme from the metagenome for further characterization. The enzyme, Cel240, exhibited close to 40% of its relative activity at low temperatures (4°C) and demonstrated endoglucanase characteristics, with a preference for cellulose substrates. Despite low sequence similarity with known enzymes, computational analysis and structural modelling confirmed its cellulase-family affiliation. Cel240 displayed activity at low temperatures and good stability at 25°C, activity at alkaline pH and increased activity in the presence of CaCl2, making it a promising candidate for detergent and washing industry applications.
© 2024 The Authors. Microbial Biotechnology published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors state no conflict of interest.
Figures




References
-
- Barocca, M. , Santos, G. , Gerday, C. & Collins, T. (2017) Biotechnological aspects of cold‐active enzymes. In: Margesin, R. (Ed.) Psychrophiles: from biodiversity to biotechnology, 2nd edition. Cham, Switzerland: Springer International Publishing, pp. 1–685.
-
- Bartolo‐Aguilar, Y. , Chávez‐Cabrera, C. , Flores‐Cotera, L.B. , Badillo‐Corona, J.A. , Oliver‐Salvador, C. & Marsch, R. (2022) The potential of cold‐shock promoters for the expression of recombinant proteins in microbes and mammalian cells. Journal of Genetic Engineering and Biotechnology, 20, 173. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

