Data-driven learning of structure augments quantitative prediction of biological responses
- PMID: 38829926
- PMCID: PMC11233023
- DOI: 10.1371/journal.pcbi.1012185
Data-driven learning of structure augments quantitative prediction of biological responses
Abstract
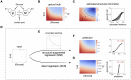
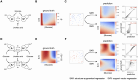
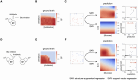
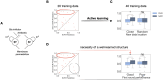
Multi-factor screenings are commonly used in diverse applications in medicine and bioengineering, including optimizing combination drug treatments and microbiome engineering. Despite the advances in high-throughput technologies, large-scale experiments typically remain prohibitively expensive. Here we introduce a machine learning platform, structure-augmented regression (SAR), that exploits the intrinsic structure of each biological system to learn a high-accuracy model with minimal data requirement. Under different environmental perturbations, each biological system exhibits a unique, structured phenotypic response. This structure can be learned based on limited data and once learned, can constrain subsequent quantitative predictions. We demonstrate that SAR requires significantly fewer data comparing to other existing machine-learning methods to achieve a high prediction accuracy, first on simulated data, then on experimental data of various systems and input dimensions. We then show how a learned structure can guide effective design of new experiments. Our approach has implications for predictive control of biological systems and an integration of machine learning prediction and experimental design.
Copyright: © 2024 Ha et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

