IL-6 inhibition prevents costimulation blockade-resistant allograft rejection in T cell-depleted recipients by promoting intragraft immune regulation in mice
- PMID: 38830846
- PMCID: PMC11148062
- DOI: 10.1038/s41467-024-48574-w
IL-6 inhibition prevents costimulation blockade-resistant allograft rejection in T cell-depleted recipients by promoting intragraft immune regulation in mice
Abstract
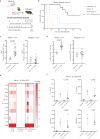
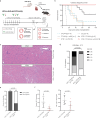
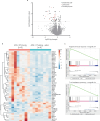
The efficacy of costimulation blockade with CTLA4-Ig (belatacept) in transplantation is limited due to T cell-mediated rejection, which also persists after induction with anti-thymocyte globulin (ATG). Here, we investigate why ATG fails to prevent costimulation blockade-resistant rejection and how this barrier can be overcome. ATG did not prevent graft rejection in a murine heart transplant model of CTLA4-Ig therapy and induced a pro-inflammatory cytokine environment. While ATG improved the balance between regulatory T cells (Treg) and effector T cells in the spleen, it had no such effect within cardiac allografts. Neutralizing IL-6 alleviated graft inflammation, increased intragraft Treg frequencies, and enhanced intragraft IL-10 and Th2-cytokine expression. IL-6 blockade together with ATG allowed CTLA4-Ig therapy to achieve long-term, rejection-free heart allograft survival. This beneficial effect was abolished upon Treg depletion. Combining ATG with IL-6 blockade prevents costimulation blockade-resistant rejection, thereby eliminating a major impediment to clinical use of costimulation blockers in transplantation.
© 2024. The Author(s).
Conflict of interest statement
TW received speaker’s honoraria from eGenesis and Mallinckrodt/Therakos and is a DSMB member for Quell Therapeutics. The other authors have no competing interests to disclose.
Figures



Similar articles
-
Complement Dependence of Murine Costimulatory Blockade-Resistant Cellular Cardiac Allograft Rejection.Am J Transplant. 2017 Nov;17(11):2810-2819. doi: 10.1111/ajt.14328. Epub 2017 May 30. Am J Transplant. 2017. PMID: 28444847 Free PMC article.
-
A novel clinically relevant approach to tip the balance toward regulation in stringent transplant model.Transplantation. 2010 Aug 15;90(3):260-9. doi: 10.1097/tp.0b013e3181e64217. Transplantation. 2010. PMID: 20712076 Free PMC article.
-
The Immunosuppressive Effect of CTLA4 Immunoglobulin Is Dependent on Regulatory T Cells at Low But Not High Doses.Am J Transplant. 2016 Dec;16(12):3404-3415. doi: 10.1111/ajt.13872. Epub 2016 Jul 5. Am J Transplant. 2016. PMID: 27184870
-
Relative roles of Th1 and Th17 effector cells in allograft rejection.Curr Opin Organ Transplant. 2009 Feb;14(1):23-9. doi: 10.1097/MOT.0b013e32831b70c2. Curr Opin Organ Transplant. 2009. PMID: 19337142 Review.
-
The potential of autologous regulatory T cell (Treg) therapy to prevent Cardiac Allograft Vasculopathy (CAV) in paediatric heart transplant recipients.Front Immunol. 2024 Sep 9;15:1444924. doi: 10.3389/fimmu.2024.1444924. eCollection 2024. Front Immunol. 2024. PMID: 39315099 Free PMC article. Review.
Cited by
-
Tocilizumab-Based Treatment of Microvascular Inflammation in Kidney Transplant Recipients: A Retrospective Study.Transpl Int. 2025 May 16;38:14502. doi: 10.3389/ti.2025.14502. eCollection 2025. Transpl Int. 2025. PMID: 40454296 Free PMC article.
-
Immunosuppression Management in Heart Transplantation.Methodist Debakey Cardiovasc J. 2025 May 15;21(3):40-50. doi: 10.14797/mdcvj.1596. eCollection 2025. Methodist Debakey Cardiovasc J. 2025. PMID: 40384742 Free PMC article. Review.
-
Novel pretransplant desensitization strategies in heart transplantation.JHLT Open. 2025 Mar 6;8:100242. doi: 10.1016/j.jhlto.2025.100242. eCollection 2025 May. JHLT Open. 2025. PMID: 40486117 Free PMC article. Review.
-
Belatacept in Kidney Transplantation: Reflecting on the Past, Shaping the Future.Transpl Int. 2025 May 20;38:14412. doi: 10.3389/ti.2025.14412. eCollection 2025. Transpl Int. 2025. PMID: 40463417 Free PMC article. Review.
-
Case Report: Successful treatment of steroid- and ruxolitinib-refractory gastrointestinal acute graft-versus-Host disease with anti-thymocyte globulin.Front Immunol. 2025 Jul 25;16:1610071. doi: 10.3389/fimmu.2025.1610071. eCollection 2025. Front Immunol. 2025. PMID: 40787448 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

