PPIA dictates NRF2 stability to promote lung cancer progression
- PMID: 38830868
- PMCID: PMC11148020
- DOI: 10.1038/s41467-024-48364-4
PPIA dictates NRF2 stability to promote lung cancer progression
Abstract
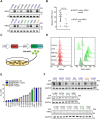
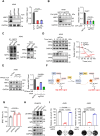
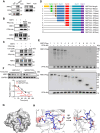
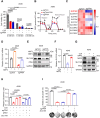
Nuclear factor erythroid 2-related factor 2 (NRF2) hyperactivation has been established as an oncogenic driver in a variety of human cancers, including non-small cell lung cancer (NSCLC). However, despite massive efforts, no specific therapy is currently available to target NRF2 hyperactivation. Here, we identify peptidylprolyl isomerase A (PPIA) is required for NRF2 protein stability. Ablation of PPIA promotes NRF2 protein degradation and blocks NRF2-driven growth in NSCLC cells. Mechanistically, PPIA physically binds to NRF2 and blocks the access of ubiquitin/Kelch Like ECH Associated Protein 1 (KEAP1) to NRF2, thus preventing ubiquitin-mediated degradation. Our X-ray co-crystal structure reveals that PPIA directly interacts with a NRF2 interdomain linker via a trans-proline 174-harboring hydrophobic sequence. We further demonstrate that an FDA-approved drug, cyclosporin A (CsA), impairs the interaction of NRF2 with PPIA, inducing NRF2 ubiquitination and degradation. Interestingly, CsA interrupts glutamine metabolism mediated by the NRF2/KLF5/SLC1A5 pathway, consequently suppressing the growth of NRF2-hyperactivated NSCLC cells. CsA and a glutaminase inhibitor combination therapy significantly retard tumor progression in NSCLC patient-derived xenograft (PDX) models with NRF2 hyperactivation. Our study demonstrates that targeting NRF2 protein stability is an actionable therapeutic approach to treat NRF2-hyperactivated NSCLC.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
PIDD interaction with KEAP1 as a new mutation-independent mechanism to promote NRF2 stabilization and chemoresistance in NSCLC.Sci Rep. 2019 Aug 27;9(1):12437. doi: 10.1038/s41598-019-48763-4. Sci Rep. 2019. PMID: 31455821 Free PMC article.
-
LKB1 and KEAP1/NRF2 Pathways Cooperatively Promote Metabolic Reprogramming with Enhanced Glutamine Dependence in KRAS-Mutant Lung Adenocarcinoma.Cancer Res. 2019 Jul 1;79(13):3251-3267. doi: 10.1158/0008-5472.CAN-18-3527. Epub 2019 Apr 30. Cancer Res. 2019. PMID: 31040157 Free PMC article.
-
E3 ligase TRIM15 facilitates non-small cell lung cancer progression through mediating Keap1-Nrf2 signaling pathway.Cell Commun Signal. 2022 May 9;20(1):62. doi: 10.1186/s12964-022-00875-7. Cell Commun Signal. 2022. PMID: 35534896 Free PMC article.
-
Role of NRF2 in Lung Cancer.Cells. 2021 Jul 24;10(8):1879. doi: 10.3390/cells10081879. Cells. 2021. PMID: 34440648 Free PMC article. Review.
-
Battles against aberrant KEAP1-NRF2 signaling in lung cancer: intertwined metabolic and immune networks.Theranostics. 2023 Jan 1;13(2):704-723. doi: 10.7150/thno.80184. eCollection 2023. Theranostics. 2023. PMID: 36632216 Free PMC article. Review.
Cited by
-
Redox-Induced Stabilization of AMBRA1 by USP7 Promotes Intestinal Oxidative Stress and Colitis Through Antagonizing DUB3-Mediated NRF2 Deubiquitination.Adv Sci (Weinh). 2025 Mar;12(12):e2411320. doi: 10.1002/advs.202411320. Epub 2025 Jan 31. Adv Sci (Weinh). 2025. PMID: 39887666 Free PMC article.
-
Target agnostic cellular screening in the era of chemically induced proximity.RSC Med Chem. 2025 Jul 21. doi: 10.1039/d5md00529a. Online ahead of print. RSC Med Chem. 2025. PMID: 40756522 Free PMC article. Review.
-
NRF2 Deficiency in Bladder Epithelial Cells Owing to Ubiquitination by N6-Methyladenosine-Modified TRIM21 Induces Oxidative Stress and Inflammation to Aggravate IC/BPS.J Inflamm Res. 2025 Aug 24;18:11577-11592. doi: 10.2147/JIR.S545880. eCollection 2025. J Inflamm Res. 2025. PMID: 40896536 Free PMC article.
-
The Crosstalk between Autophagy and Nrf2 Signaling in Cancer: from Biology to Clinical Applications.Int J Biol Sci. 2024 Nov 11;20(15):6181-6206. doi: 10.7150/ijbs.103187. eCollection 2024. Int J Biol Sci. 2024. PMID: 39664581 Free PMC article. Review.
-
Tumor microenvironment regulation by reactive oxygen species-mediated inflammasome activation.Arch Pharm Res. 2025 Feb;48(2):115-131. doi: 10.1007/s12272-025-01532-6. Epub 2025 Jan 31. Arch Pharm Res. 2025. PMID: 39888519 Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

