O-GlcNAcylation controls pro-fibrotic transcriptional regulatory signaling in myofibroblasts
- PMID: 38830870
- PMCID: PMC11148087
- DOI: 10.1038/s41419-024-06773-9
O-GlcNAcylation controls pro-fibrotic transcriptional regulatory signaling in myofibroblasts
Abstract
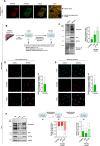
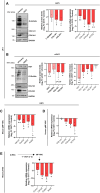
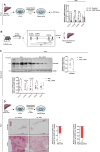
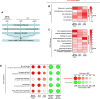
Tissue injury causes activation of mesenchymal lineage cells into wound-repairing myofibroblasts (MFs), whose uncontrolled activity ultimately leads to fibrosis. Although this process is triggered by deep metabolic and transcriptional reprogramming, functional links between these two key events are not yet understood. Here, we report that the metabolic sensor post-translational modification O-linked β-D-N-acetylglucosaminylation (O-GlcNAcylation) is increased and required for myofibroblastic activation. Inhibition of protein O-GlcNAcylation impairs archetypal myofibloblast cellular activities including extracellular matrix gene expression and collagen secretion/deposition as defined in vitro and using ex vivo and in vivo murine liver injury models. Mechanistically, a multi-omics approach combining proteomic, epigenomic, and transcriptomic data mining revealed that O-GlcNAcylation controls the MF transcriptional program by targeting the transcription factors Basonuclin 2 (BNC2) and TEA domain transcription factor 4 (TEAD4) together with the Yes-associated protein 1 (YAP1) co-activator. Indeed, inhibition of protein O-GlcNAcylation impedes their stability leading to decreased functionality of the BNC2/TEAD4/YAP1 complex towards promoting activation of the MF transcriptional regulatory landscape. We found that this involves O-GlcNAcylation of BNC2 at Thr455 and Ser490 and of TEAD4 at Ser69 and Ser99. Altogether, this study unravels protein O-GlcNAcylation as a key determinant of myofibroblastic activation and identifies its inhibition as an avenue to intervene with fibrogenic processes.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
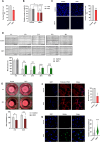

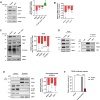
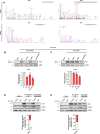
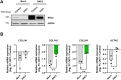
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- ANR-10-INBS-08-03/Agence Nationale de la Recherche (French National Research Agency)
- ANR-10-INBS-08-03/Agence Nationale de la Recherche (French National Research Agency)
- ANR 21 CE17-0016-02/Agence Nationale de la Recherche (French National Research Agency)
- ANR-16-RHUS-0006-PreciNASH/Agence Nationale de la Recherche (French National Research Agency)
- ANR 10 LABX-0046/Agence Nationale de la Recherche (French National Research Agency)
- ANR-16-IDEX-0004 ULNE/Agence Nationale de la Recherche (French National Research Agency)
- ANR-16-RHUS-0006-PreciNASH/Agence Nationale de la Recherche (French National Research Agency)
- ANR 10 LABX-0046/Agence Nationale de la Recherche (French National Research Agency)
- ANR-16-IDEX-0004 ULNE/Agence Nationale de la Recherche (French National Research Agency)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

