A multispectral 3D live organoid imaging platform to screen probes for fluorescence guided surgery
- PMID: 38831131
- PMCID: PMC11251264
- DOI: 10.1038/s44321-024-00084-4
A multispectral 3D live organoid imaging platform to screen probes for fluorescence guided surgery
Abstract
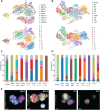
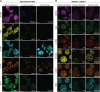
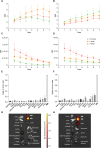
Achieving complete tumor resection is challenging and can be improved by real-time fluorescence-guided surgery with molecular-targeted probes. However, pre-clinical identification and validation of probes presents a lengthy process that is traditionally performed in animal models and further hampered by inter- and intra-tumoral heterogeneity in target expression. To screen multiple probes at patient scale, we developed a multispectral real-time 3D imaging platform that implements organoid technology to effectively model patient tumor heterogeneity and, importantly, healthy human tissue binding.
Keywords: Breast Cancer; Fluorescence-guided Surgery; Multi-spectral 3D Imaging; Neuroblastoma; Patient-derived Organoids.
© 2024. The Author(s).
Figures





References
-
- Bellacosa A, Godwin AK, Peri S, Devarajan K, Caretti E, Vanderveer L, Bove B, Slater C, Zhou Y, Daly M, et al. Altered gene expression in morphologically normal epithelial cells from heterozygous carriers of BRCA1 or BRCA2 mutations. Cancer Prev Res. 2010;3:48–61. doi: 10.1158/1940-6207.CAPR-09-0078. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

