Atg45 is an autophagy receptor for glycogen, a non-preferred cargo of bulk autophagy in yeast
- PMID: 38832010
- PMCID: PMC11145338
- DOI: 10.1016/j.isci.2024.109810
Atg45 is an autophagy receptor for glycogen, a non-preferred cargo of bulk autophagy in yeast
Abstract
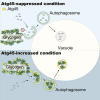
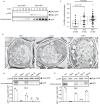
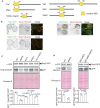
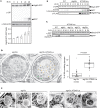
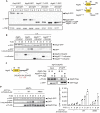
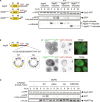
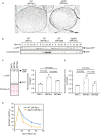
The mechanisms governing autophagy of proteins and organelles have been well studied, but how other cytoplasmic components such as RNA and polysaccharides are degraded remains largely unknown. In this study, we examine autophagy of glycogen, a storage form of glucose. We find that cells accumulate glycogen in the cytoplasm during nitrogen starvation and that this carbohydrate is rarely observed within autophagosomes and autophagic bodies. However, sequestration of glycogen by autophagy is observed following prolonged nitrogen starvation. We identify a yet-uncharacterized open reading frame, Yil024c (herein Atg45), as encoding a cytosolic receptor protein that mediates autophagy of glycogen (glycophagy). Furthermore, we show that, during sporulation, Atg45 is highly expressed and is associated with an increase in glycophagy. Our results suggest that cells regulate glycophagic activity by controlling the expression level of Atg45.
Keywords: Biomolecules; Cell biology; Glycobiology; Model organism.
© 2024 The Authors.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

