Exosomal lncRNA SNHG12 promotes angiogenesis and breast cancer progression
- PMID: 38833118
- PMCID: PMC11194216
- DOI: 10.1007/s12282-024-01574-6
Exosomal lncRNA SNHG12 promotes angiogenesis and breast cancer progression
Abstract
Objective: Breast cancer is one of the most prevalent malignancies in women. Exosomes are important mediators of intercellular communication; however, their regulatory mechanisms in human umbilical vein endothelial cells (HUVECs) angiogenesis in breast cancer remain unknown.
Methods: We isolated and characterized breast cancer cell-derived exosomes and investigated their functions. Exosomal sequencing and the TCGA database were used to screen long non-coding RNA (lncRNA). In vitro and in vivo experiments were performed to investigate the role of exosomal lncRNA in HUVEC angiogenesis and tumor growth. Molecular methods were used to demonstrate the molecular mechanism of lncRNA.
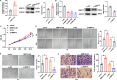
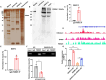
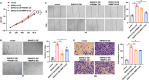
Results: We demonstrated that breast cancer cell-derived exosomes promoted HUVEC proliferation, tube formation, and migration. Combining exosomal sequencing results with The Cancer Genome Atlas Breast Cancer database, we screened lncRNA small nucleolar RNA host gene 12 (SNHG12), which was highly expressed in breast cancer cells. SNHG12 was also upregulated in HUVECs co-cultured with exosome-overexpressed SNHG12. Moreover, overexpression of SNHG12 in exosomes increased HUVEC proliferation and migration, whereas deletion of SNHG12 in exosomes showed the opposite effects. In vivo experiments showed that SNHG12 knockdown in exosomes inhibited breast cancer tumor growth. Transcriptome sequencing identified MMP10 as the target gene of SNHG12. Functional experiments revealed that MMP10 overexpression promoted HUVEC angiogenesis. Mechanistically, SNHG12 blocked the interaction between PBRM1 and MMP10 by directly binding to PBRM1. Moreover, exosomal SNHG12 promoted HUVEC angiogenesis via PBRM1 and MMP10.
Conclusions: In summary, our findings confirmed that exosomal SNHG12 promoted HUVEC angiogenesis via the PBRM1-MMP10 axis, leading to enhanced malignancy of breast cancer. Exosomal SNHG12 may be a novel therapeutic target for breast cancer.
Keywords: Angiogenesis; Breast cancer; Exosomes; Progression; lncRNA SNHG12.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures

Similar articles
-
Melanoma cell line-derived exosomal miR-424-5p: a key promoter of angiogenesis through LATS2 interaction.Oncol Res. 2025 Jan 16;33(2):357-367. doi: 10.32604/or.2024.050878. eCollection 2025. Oncol Res. 2025. PMID: 39866229 Free PMC article.
-
[Long non-coding RNA colon cancer associated transcript-2 from nasopharyngeal carcinoma-derived exosomes promotes angiogenesis].Zhonghua Er Bi Yan Hou Tou Jing Wai Ke Za Zhi. 2020 Oct 7;55(10):944-951. doi: 10.3760/cma.j.cn115330-20200423-00322. Zhonghua Er Bi Yan Hou Tou Jing Wai Ke Za Zhi. 2020. PMID: 33036509 Chinese.
-
MEG3-Mediated Oral Squamous-Cell-Carcinoma-Derived Exosomal miR-421 Activates Angiogenesis by Targeting HS2ST1 in Vascular Endothelial Cells.Int J Mol Sci. 2024 Jul 10;25(14):7576. doi: 10.3390/ijms25147576. Int J Mol Sci. 2024. PMID: 39062818 Free PMC article.
-
Exosomal lncRNAs in the Tumor Angiogenesis: As Therapeutic Targets in Cancer Treatment.Arch Pharm (Weinheim). 2025 Apr;358(4):e202400940. doi: 10.1002/ardp.202400940. Arch Pharm (Weinheim). 2025. PMID: 40165644 Review.
-
Potential biological roles of exosomal non-coding RNAs in breast cancer.FASEB J. 2025 Mar 31;39(6):e70456. doi: 10.1096/fj.202500022R. FASEB J. 2025. PMID: 40079186 Free PMC article. Review.
Cited by
-
Latest Update on lncRNA in Epithelial Ovarian Cancer-A Scoping Review.Cells. 2025 Apr 7;14(7):555. doi: 10.3390/cells14070555. Cells. 2025. PMID: 40214508 Free PMC article.
-
Exosomal long non-coding RNAs in human malignancies: biological functions and clinical applications.Med Oncol. 2025 Jun 24;42(8):279. doi: 10.1007/s12032-025-02845-8. Med Oncol. 2025. PMID: 40553231 Review.
-
Development of a coagulation‑related gene model for prognostication, immune response and treatment prediction in lung adenocarcinoma.Oncol Lett. 2025 Apr 11;29(6):290. doi: 10.3892/ol.2025.15035. eCollection 2025 Jun. Oncol Lett. 2025. PMID: 40276086 Free PMC article.
-
LINC01559: roles, mechanisms, and clinical implications in human cancers.Hum Cell. 2025 Apr 9;38(3):83. doi: 10.1007/s13577-025-01218-7. Hum Cell. 2025. PMID: 40205068 Review.
-
Investigating the role of exosomal long non-coding RNAs in drug resistance within female reproductive system cancers.Front Cell Dev Biol. 2025 Jan 24;13:1485422. doi: 10.3389/fcell.2025.1485422. eCollection 2025. Front Cell Dev Biol. 2025. PMID: 39925739 Free PMC article. Review.
References
-
- Peart O. Breast intervention and breast cancer treatment options. Radiol Technol. 2015;86:535M–58M; quiz 59–62. - PubMed
-
- Global Burden of Disease Cancer Collaboration Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 29 cancer groups, 1990–2016: a systematic analysis for the Global Burden of Disease Study. JAMA Oncol. 2018;4:1553–1568. doi: 10.1001/jamaoncol.2018.2706. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

