BioInstruct: instruction tuning of large language models for biomedical natural language processing
- PMID: 38833265
- PMCID: PMC11339494
- DOI: 10.1093/jamia/ocae122
BioInstruct: instruction tuning of large language models for biomedical natural language processing
Abstract
Objectives: To enhance the performance of large language models (LLMs) in biomedical natural language processing (BioNLP) by introducing a domain-specific instruction dataset and examining its impact when combined with multi-task learning principles.
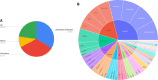
Materials and methods: We created the BioInstruct, comprising 25 005 instructions to instruction-tune LLMs (LLaMA 1 and 2, 7B and 13B version). The instructions were created by prompting the GPT-4 language model with 3-seed samples randomly drawn from an 80 human curated instructions. We employed Low-Rank Adaptation (LoRA) for parameter-efficient fine-tuning. We then evaluated these instruction-tuned LLMs on several BioNLP tasks, which can be grouped into 3 major categories: question answering (QA), information extraction (IE), and text generation (GEN). We also examined whether categories (eg, QA, IE, and generation) of instructions impact model performance.
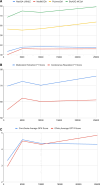
Results and discussion: Comparing with LLMs without instruction-tuned, our instruction-tuned LLMs demonstrated marked performance gains: 17.3% in QA on average accuracy metric, 5.7% in IE on average F1 metric, and 96% in Generation tasks on average GPT-4 score metric. Our 7B-parameter instruction-tuned LLaMA 1 model was competitive or even surpassed other LLMs in the biomedical domain that were also fine-tuned from LLaMA 1 with vast domain-specific data or a variety of tasks. Our results also show that the performance gain is significantly higher when instruction fine-tuning is conducted with closely related tasks. Our findings align with the observations of multi-task learning, suggesting the synergies between 2 tasks.
Conclusion: The BioInstruct dataset serves as a valuable resource and instruction tuned LLMs lead to the best performing BioNLP applications.
Keywords: information extraction; instruction tuning; large language models; multi-task learning; natural language inference; question answering; text generation.
© The Author(s) 2024. Published by Oxford University Press on behalf of the American Medical Informatics Association. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Conflict of interest statement
The authors declare no competing interests.
Figures
References
-
- Brown TB, Mann B, Ryder N, et al. Language models are few-shot learners. In: Larochelle H, Ranzato M, Hadsell R, Balcan MF, Lin H, eds. Advances in Neural Information Processing Systems. Vol 33. 2020:1877-1901. https://proceedings.neurips.cc/paper/2020/file/1457c0d6bfcb4967418bfb8ac....
-
- Sanh V, Webson A, Raffel C, et al. 2021. Multitask prompted training enables zero-shot task generalization. CoRR; abs/2110.08207. https://arxiv.org/abs/2110.08207.
-
- Chowdhery A, Narang S, Devlin J, et al. 2022. PaLM: scaling language modeling with pathways. arXiv, arXiv:220402311, preprint: not peer reviewed. https://arxiv.org/abs/2204.02311.
-
- Longpre S, Hou L, Vu T, et al. 2023. The flan collection: designing data and methods for effective instruction tuning. https://arxiv.org/abs/2301.13688.
-
- OpenAI. 2023. GPT-4 Technical Report. arXiv, arXiv:230308774, preprint: not peer reviewed. https://api.semanticscholar.org/CorpusID:257532815.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

