Effects of Developmental Lead and Phthalate Exposures on DNA Methylation in Adult Mouse Blood, Brain, and Liver: A Focus on Genomic Imprinting by Tissue and Sex
- PMID: 38833407
- PMCID: PMC11166413
- DOI: 10.1289/EHP14074
Effects of Developmental Lead and Phthalate Exposures on DNA Methylation in Adult Mouse Blood, Brain, and Liver: A Focus on Genomic Imprinting by Tissue and Sex
Abstract
Background: Maternal exposure to environmental chemicals can cause adverse health effects in offspring. Mounting evidence supports that these effects are influenced, at least in part, by epigenetic modifications. It is unknown whether epigenetic changes in surrogate tissues such as the blood are reflective of similar changes in target tissues such as cortex or liver.
Objective: We examined tissue- and sex-specific changes in DNA methylation (DNAm) associated with human-relevant lead (Pb) and di(2-ethylhexyl) phthalate (DEHP) exposure during perinatal development in cerebral cortex, blood, and liver.
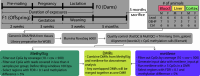
Methods: Female mice were exposed to human relevant doses of either Pb () via drinking water or DEHP () via chow for 2 weeks prior to mating through offspring weaning. Whole genome bisulfite sequencing (WGBS) was utilized to examine DNAm changes in offspring cortex, blood, and liver at 5 months of age. Metilene and methylSig were used to identify differentially methylated regions (DMRs). Annotatr and ChIP-enrich were used for genomic annotations and gene set enrichment tests of DMRs, respectively.
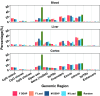
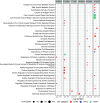
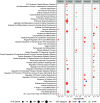
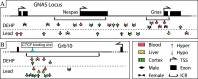
Results: The cortex contained the majority of DMRs associated with Pb (66%) and DEHP (57%) exposure. The cortex also contained the greatest degree of overlap in DMR signatures between sexes ( and 8 DMRs with Pb and DEHP exposure, respectively) and exposure types ( and 39 DMRs in males and females, respectively). In all tissues, detected DMRs were preferentially found at genomic regions associated with gene expression regulation (e.g., CpG islands and shores, 5' UTRs, promoters, and exons). An analysis of GO terms associated with DMR-containing genes identified imprinted genes to be impacted by both Pb and DEHP exposure. Of these, Gnas and Grb10 contained DMRs across tissues, sexes, and exposures, with some signatures replicated between target and surrogate tissues. DMRs were enriched in the imprinting control regions (ICRs) of Gnas and Grb10, and we again observed a replication of DMR signatures between blood and target tissues. Specifically, we observed hypermethylation of the Grb10 ICR in both blood and liver of Pb-exposed male animals.
Conclusions: These data provide preliminary evidence that imprinted genes may be viable candidates in the search for epigenetic biomarkers of toxicant exposure in target tissues. Additional research is needed on allele- and developmental stage-specific effects, as well as whether other imprinted genes provide additional examples of this relationship. https://doi.org/10.1289/EHP14074.
Figures


Update of
-
Effects of Developmental Lead and Phthalate Exposures on DNA Methylation in Adult Mouse Blood, Brain, and Liver Identifies Tissue- and Sex-Specific Changes with Implications for Genomic Imprinting.bioRxiv [Preprint]. 2023 Oct 10:2023.09.29.560131. doi: 10.1101/2023.09.29.560131. bioRxiv. 2023. Update in: Environ Health Perspect. 2024 Jun;132(6):67003. doi: 10.1289/EHP14074. PMID: 37873115 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
- K01 ES032048/ES/NIEHS NIH HHS/United States
- P30 CA046592/CA/NCI NIH HHS/United States
- R01 AG072396/AG/NIA NIH HHS/United States
- P30 ES013508/ES/NIEHS NIH HHS/United States
- P30 ES017885/ES/NIEHS NIH HHS/United States
- P30 DK020572/DK/NIDDK NIH HHS/United States
- U01 ES026697/ES/NIEHS NIH HHS/United States
- K99 ES034429/ES/NIEHS NIH HHS/United States
- R01 ES007062/ES/NIEHS NIH HHS/United States
- K01 ES035064/ES/NIEHS NIH HHS/United States
- K99 HD112543/HD/NICHD NIH HHS/United States
- T32 HD079342/HD/NICHD NIH HHS/United States
- R01 ES028802/ES/NIEHS NIH HHS/United States
- T32 ES007062/ES/NIEHS NIH HHS/United States
- R35 ES031686/ES/NIEHS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Miscellaneous
