Multivariate genetic architecture reveals testosterone-driven sexual antagonism in contemporary humans
- PMID: 38833469
- PMCID: PMC11181031
- DOI: 10.1073/pnas.2404364121
Multivariate genetic architecture reveals testosterone-driven sexual antagonism in contemporary humans
Abstract
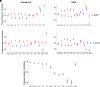
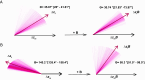
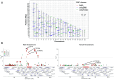
Sex difference (SD) is ubiquitous in humans despite shared genetic architecture (SGA) between the sexes. A univariate approach, i.e., studying SD in single traits by estimating genetic correlation, does not provide a complete biological overview, because traits are not independent and are genetically correlated. The multivariate genetic architecture between the sexes can be summarized by estimating the additive genetic (co)variance across shared traits, which, apart from the cross-trait and cross-sex covariances, also includes the cross-sex-cross-trait covariances, e.g., between height in males and weight in females. Using such a multivariate approach, we investigated SD in the genetic architecture of 12 anthropometric, fat depositional, and sex-hormonal phenotypes. We uncovered sexual antagonism (SA) in the cross-sex-cross-trait covariances in humans, most prominently between testosterone and the anthropometric traits - a trend similar to phenotypic correlations. 27% of such cross-sex-cross-trait covariances were of opposite sign, contributing to asymmetry in the SGA. Intriguingly, using multivariate evolutionary simulations, we observed that the SGA acts as a genetic constraint to the evolution of SD in humans only when selection is sexually antagonistic and not concordant. Remarkably, we found that the lifetime reproductive success in both the sexes shows a positive genetic correlation with anthropometric traits, but not with testosterone. Moreover, we demonstrated that genetic variance is depleted along multivariate trait combinations in both the sexes but in different directions, suggesting absolute genetic constraint to evolution. Our results indicate that testosterone drives SA in contemporary humans and emphasize the necessity and significance of using a multivariate framework in studying SD.
Keywords: B matrix; additive genetic (co)variance matrix; genetic correlations; human complex traits; sex difference.
Conflict of interest statement
Competing interests statement:S.C. is currently an employee of GSK. The work included in this manuscript was performed when S.C. was working at the Biotechnology Research Innovation Council- National Institute of Biomedical Genomics, Kalyani. All views and opinions expressed in this manuscript are solely of the authors’ and do not in any manner reflect the official stand of GSK.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

