BATF-dependent Th17 cells act through the IL-23R pathway to promote prostate adenocarcinoma initiation and progression
- PMID: 38833676
- PMCID: PMC11461145
- DOI: 10.1093/jnci/djae120
BATF-dependent Th17 cells act through the IL-23R pathway to promote prostate adenocarcinoma initiation and progression
Abstract
Background: The role of Th17 cells in prostate cancer is not fully understood. The transcription factor BATF controls the differentiation of Th17 cells. Mice deficient in Batf do not produce Th17 cells.
Methods: In this study, we aimed to characterize the role of Batf-dependent Th17 cells in prostate cancer by crossbreeding Batf knockout mice with mice conditionally mutant for Pten.
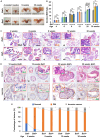
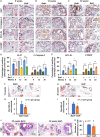
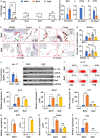
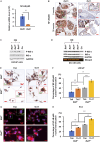
Results: We found that Batf knockout mice had changes in the morphology of prostate epithelial cells compared with normal mice, and Batf knockout mice deficient in Pten (called Batf-) had smaller prostate size and developed fewer invasive prostate adenocarcinomas than Pten-deficient mice with Batf expression (called Batf+). The prostate tumors in Batf- mice showed reduced proliferation, increased apoptosis, decreased angiogenesis and inflammatory cell infiltration, and activation of nuclear factor-κB signaling. Moreover, Batf- mice showed significantly reduced interleukin 23 (IL-23)-IL-23R signaling. In the prostate stroma of Batf- mice, IL-23R-positive cells were decreased considerably compared with Batf+ mice. Splenocytes and prostate tissues from Batf- mice cultured under Th17 differentiation conditions expressed reduced IL-23/IL-23R than cultured cells from Batf+ mice. Anti-IL-23p19 antibody treatment of Pten-deficient mice reduced prostate tumors and angiogenesis compared with control immunoglobulin G-treated mice. In human prostate tumors, BATF messenger RNA level was positively correlated with IL-23A and IL-23R but not RORC.
Conclusion: Our novel findings underscore the crucial role of IL-23-IL-23R signaling in mediating the function of Batf-dependent Th17 cells, thereby promoting prostate cancer initiation and progression. This finding highlights the BATF-IL-23R axis as a promising target for the development of innovative strategies for prostate cancer prevention and treatment.
© The Author(s) 2024. Published by Oxford University Press.
Conflict of interest statement
The authors declare no potential conflicts of interest.
Figures



Similar articles
-
atRA Attenuates High Salt-Driven EAE Mainly Through Suppressing Th17-Like Regulatory T Cell Response Mediated by the Inhibition of IL-23R Signaling Pathway.Inflammation. 2025 Jun;48(3):1403-1419. doi: 10.1007/s10753-024-02130-2. Epub 2024 Aug 21. Inflammation. 2025. PMID: 39167321
-
Crosstalk Between Th17 Cells and Renal Tubular Epithelial Cells Promotes Fibrotic Progression in IgA Nephropathy.Curr Med Sci. 2025 Jun;45(3):626-639. doi: 10.1007/s11596-025-00068-6. Epub 2025 Jun 11. Curr Med Sci. 2025. PMID: 40498423
-
Batf-dependent Th17 cells critically regulate IL-23 driven colitis-associated colon cancer.Gut. 2016 Jul;65(7):1139-50. doi: 10.1136/gutjnl-2014-308227. Epub 2015 Apr 2. Gut. 2016. PMID: 25838550
-
Impact of residual disease as a prognostic factor for survival in women with advanced epithelial ovarian cancer after primary surgery.Cochrane Database Syst Rev. 2022 Sep 26;9(9):CD015048. doi: 10.1002/14651858.CD015048.pub2. Cochrane Database Syst Rev. 2022. PMID: 36161421 Free PMC article.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
Cited by
-
Prostate Cancer Bone Metastasis: Molecular Mechanisms of Tumor and Bone Microenvironment.Cancer Manag Res. 2025 Feb 1;17:219-237. doi: 10.2147/CMAR.S495169. eCollection 2025. Cancer Manag Res. 2025. PMID: 39912095 Free PMC article. Review.
-
The RNA binding protein CARHSP1 facilitates tumor growth, metastasis and immune escape by enhancing IL-17RA mRNA stabilization in prostate cancer.Cell Biosci. 2025 Mar 7;15(1):33. doi: 10.1186/s13578-025-01371-4. Cell Biosci. 2025. PMID: 40055805 Free PMC article.
-
IL-17A in gastric carcinogenesis: good or bad?Front Immunol. 2024 Nov 29;15:1501293. doi: 10.3389/fimmu.2024.1501293. eCollection 2024. Front Immunol. 2024. PMID: 39676857 Free PMC article. Review.
-
The CCL5/CCR5/SHP2 axis sustains Stat1 phosphorylation and activates NF-κB signaling promoting M1 macrophage polarization and exacerbating chronic prostatic inflammation.Cell Commun Signal. 2024 Dec 5;22(1):584. doi: 10.1186/s12964-024-01943-w. Cell Commun Signal. 2024. PMID: 39633456 Free PMC article.
References
-
- Giaquinto AN, Sung H, Miller KD, et al.Cancer statistics, 2022. CA Cancer J Clin. 2022;72(6):524-541. - PubMed
-
- Schatteman PH, Hoekx L, Wyndaele JJ, Jeuris W, Van Marck E.. Inflammation in prostate biopsies of men without prostatic malignancy or clinical prostatitis: correlation with total serum PSA and PSA density. Eur Urol. 2000;37(4):404-412. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

