Foundation models in ophthalmology
- PMID: 38834291
- PMCID: PMC11503093
- DOI: 10.1136/bjo-2024-325459
Foundation models in ophthalmology
Abstract
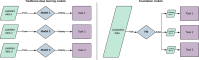
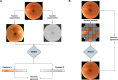
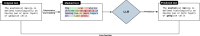
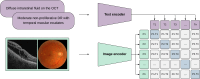
Foundation models represent a paradigm shift in artificial intelligence (AI), evolving from narrow models designed for specific tasks to versatile, generalisable models adaptable to a myriad of diverse applications. Ophthalmology as a specialty has the potential to act as an exemplar for other medical specialties, offering a blueprint for integrating foundation models broadly into clinical practice. This review hopes to serve as a roadmap for eyecare professionals seeking to better understand foundation models, while equipping readers with the tools to explore the use of foundation models in their own research and practice. We begin by outlining the key concepts and technological advances which have enabled the development of these models, providing an overview of novel training approaches and modern AI architectures. Next, we summarise existing literature on the topic of foundation models in ophthalmology, encompassing progress in vision foundation models, large language models and large multimodal models. Finally, we outline major challenges relating to privacy, bias and clinical validation, and propose key steps forward to maximise the benefit of this powerful technology.
Keywords: Imaging; Retina.
© Author(s) (or their employer(s)) 2024. Re-use permitted under CC BY. Published by BMJ.
Conflict of interest statement
Competing interests: PAK has acted as a consultant for DeepMind, Roche, Novartis, Apellis and BitFount and is an equity owner in Big Picture Medical. He has received speaker fees from Heidelberg Engineering, Topcon, Allergan and Bayer. AYL reports grants from Santen, personal fees from Genentech, personal fees from US FDA, personal fees from Johnson and Johnson, personal fees from Boehringer Ingelheim, non-financial support from iCareWorld, grants from Topcon, grants from Carl Zeiss Meditec, personal fees from Gyroscope, non-financial support from Optomed, non-financial support from Heidelberg, non-financial support from Microsoft, grants from Regeneron, grants from Amazon, grants from Meta, outside the submitted work; this article does not reflect the views of the US FDA.
Figures

Similar articles
-
AI image generation technology in ophthalmology: Use, misuse and future applications.Prog Retin Eye Res. 2025 May;106:101353. doi: 10.1016/j.preteyeres.2025.101353. Epub 2025 Mar 17. Prog Retin Eye Res. 2025. PMID: 40107410 Review.
-
Revolutionizing Digital Pathology With the Power of Generative Artificial Intelligence and Foundation Models.Lab Invest. 2023 Nov;103(11):100255. doi: 10.1016/j.labinv.2023.100255. Epub 2023 Sep 26. Lab Invest. 2023. PMID: 37757969 Review.
-
Novel technical and privacy-preserving technology for artificial intelligence in ophthalmology.Curr Opin Ophthalmol. 2022 May 1;33(3):174-187. doi: 10.1097/ICU.0000000000000846. Epub 2022 Mar 9. Curr Opin Ophthalmol. 2022. PMID: 35266894 Review.
-
Gemini AI vs. ChatGPT: A comprehensive examination alongside ophthalmology residents in medical knowledge.Graefes Arch Clin Exp Ophthalmol. 2025 Feb;263(2):527-536. doi: 10.1007/s00417-024-06625-4. Epub 2024 Sep 15. Graefes Arch Clin Exp Ophthalmol. 2025. PMID: 39277830
-
A review of ophthalmology education in the era of generative artificial intelligence.Asia Pac J Ophthalmol (Phila). 2024 Jul-Aug;13(4):100089. doi: 10.1016/j.apjo.2024.100089. Epub 2024 Aug 10. Asia Pac J Ophthalmol (Phila). 2024. PMID: 39134176 Free PMC article. Review.
Cited by
-
Foundation models in ophthalmology: opportunities and challenges.Curr Opin Ophthalmol. 2025 Jan 1;36(1):90-98. doi: 10.1097/ICU.0000000000001091. Epub 2024 Nov 4. Curr Opin Ophthalmol. 2025. PMID: 39329204 Free PMC article. Review.
-
Re-identification of patients from imaging features extracted by foundation models.NPJ Digit Med. 2025 Jul 22;8(1):469. doi: 10.1038/s41746-025-01801-0. NPJ Digit Med. 2025. PMID: 40696111 Free PMC article.
-
Automated Identification of Stroke Thrombolysis Contraindications from Synthetic Clinical Notes: A Proof-of-Concept Study.Cerebrovasc Dis Extra. 2025;15(1):130-136. doi: 10.1159/000545317. Epub 2025 Mar 17. Cerebrovasc Dis Extra. 2025. PMID: 40096831 Free PMC article.
-
Large Language Models in Ophthalmology: A Review of Publications from Top Ophthalmology Journals.Ophthalmol Sci. 2024 Dec 17;5(3):100681. doi: 10.1016/j.xops.2024.100681. eCollection 2025 May-Jun. Ophthalmol Sci. 2024. PMID: 40114712 Free PMC article.
-
Fast TILs-A pipeline for efficient TILs estimation in non-small cell Lung cancer.J Pathol Inform. 2025 Mar 12;17:100437. doi: 10.1016/j.jpi.2025.100437. eCollection 2025 Apr. J Pathol Inform. 2025. PMID: 40230809 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
