DYRK1B blockade promotes tumoricidal macrophage activity in pancreatic cancer
- PMID: 38834297
- PMCID: PMC11420735
- DOI: 10.1136/gutjnl-2023-331854
DYRK1B blockade promotes tumoricidal macrophage activity in pancreatic cancer
Abstract
Objective: Highly malignant pancreatic ductal adenocarcinoma (PDAC) is characterised by an abundant immunosuppressive and fibrotic tumour microenvironment (TME). Future therapeutic attempts will therefore demand the targeting of tumours and stromal compartments in order to be effective. Here we investigate whether dual specificity and tyrosine phosphorylation-regulated kinase 1B (DYRK1B) fulfil these criteria and represent a promising anticancer target in PDAC.
Design: We used transplantation and autochthonous mouse models of PDAC with either genetic Dyrk1b loss or pharmacological DYRK1B inhibition, respectively. Mechanistic interactions between tumour cells and macrophages were studied in direct or indirect co-culture experiments. Histological analyses used tissue microarrays from patients with PDAC. Additional methodological approaches included bulk mRNA sequencing (transcriptomics) and proteomics (secretomics).
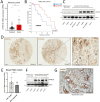
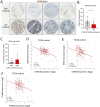
Results: We found that DYRK1B is mainly expressed by pancreatic epithelial cancer cells and modulates the influx and activity of TME-associated macrophages through effects on the cancer cells themselves as well as through the tumour secretome. Mechanistically, genetic ablation or pharmacological inhibition of DYRK1B strongly attracts tumoricidal macrophages and, in addition, downregulates the phagocytosis checkpoint and 'don't eat me' signal CD24 on cancer cells, resulting in enhanced tumour cell phagocytosis. Consequently, tumour cells lacking DYRK1B hardly expand in transplantation experiments, despite their rapid growth in culture. Furthermore, combining a small-molecule DYRK1B-directed therapy with mammalian target of rapamycin inhibition and conventional chemotherapy stalls the growth of established tumours and results in a significant extension of life span in a highly aggressive autochthonous model of PDAC.
Conclusion: In light of DYRK inhibitors currently entering clinical phase testing, our data thus provide a novel and clinically translatable approach targeting both the cancer cell compartment and its microenvironment.
Keywords: immunotherapy; macrophages; molecular carcinogenesis; pancreatic cancer; pancreatic tumours.
© Author(s) (or their employer(s)) 2024. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: None declared.
Figures





References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
