Semaglutide ameliorates cardiac remodeling in male mice by optimizing energy substrate utilization through the Creb5/NR4a1 axis
- PMID: 38834564
- PMCID: PMC11150406
- DOI: 10.1038/s41467-024-48970-2
Semaglutide ameliorates cardiac remodeling in male mice by optimizing energy substrate utilization through the Creb5/NR4a1 axis
Abstract
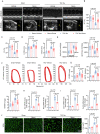
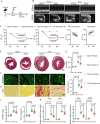
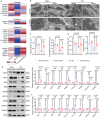
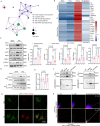
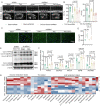
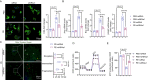
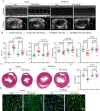
Semaglutide, a glucagon-like peptide-1 receptor agonist, is clinically used as a glucose-lowering and weight loss medication due to its effects on energy metabolism. In heart failure, energy production is impaired due to altered mitochondrial function and increased glycolysis. However, the impact of semaglutide on cardiomyocyte metabolism under pressure overload remains unclear. Here we demonstrate that semaglutide improves cardiac function and reduces hypertrophy and fibrosis in a mouse model of pressure overload-induced heart failure. Semaglutide preserves mitochondrial structure and function under chronic stress. Metabolomics reveals that semaglutide reduces mitochondrial damage, lipid accumulation, and ATP deficiency by promoting pyruvate entry into the tricarboxylic acid cycle and increasing fatty acid oxidation. Transcriptional analysis shows that semaglutide regulates myocardial energy metabolism through the Creb5/NR4a1 axis in the PI3K/AKT pathway, reducing NR4a1 expression and its translocation to mitochondria. NR4a1 knockdown ameliorates mitochondrial dysfunction and abnormal glucose and lipid metabolism in the heart. These findings suggest that semaglutide may be a therapeutic agent for improving cardiac remodeling by modulating energy metabolism.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

