Transcriptional profiling links unique human macrophage phenotypes to the growth of intracellular Salmonella enterica serovar Typhi
- PMID: 38834738
- PMCID: PMC11150401
- DOI: 10.1038/s41598-024-63588-6
Transcriptional profiling links unique human macrophage phenotypes to the growth of intracellular Salmonella enterica serovar Typhi
Abstract
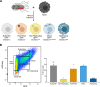
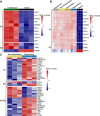
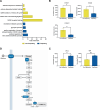
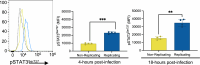
Macrophages provide a crucial environment for Salmonella enterica serovar Typhi (S. Typhi) to multiply during typhoid fever, yet our understanding of how human macrophages and S. Typhi interact remains limited. In this study, we delve into the dynamics of S. Typhi replication within human macrophages and the resulting heterogeneous transcriptomic responses of macrophages during infection. Our study reveals key factors that influence macrophage diversity, uncovering distinct immune and metabolic pathways associated with different stages of S. Typhi intracellular replication in macrophages. Of note, we found that macrophages harboring replicating S. Typhi are skewed towards an M1 pro-inflammatory state, whereas macrophages containing non-replicating S. Typhi exhibit neither a distinct M1 pro-inflammatory nor M2 anti-inflammatory state. Additionally, macrophages with replicating S. Typhi were characterized by the increased expression of genes associated with STAT3 phosphorylation and the activation of the STAT3 transcription factor. Our results shed light on transcriptomic pathways involved in the susceptibility of human macrophages to intracellular S. Typhi replication, thereby providing crucial insight into host phenotypes that restrict and support S. Typhi infection.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Salmonella enterica Serovar Typhi Induces Host Metabolic Reprogramming to Increase Glucose Availability for Intracellular Replication.Int J Mol Sci. 2021 Sep 16;22(18):10003. doi: 10.3390/ijms221810003. Int J Mol Sci. 2021. PMID: 34576166 Free PMC article.
-
Salmonella enterica serovar Typhi uses two type 3 secretion systems to replicate in human macrophages and colonize humanized mice.mBio. 2023 Aug 31;14(4):e0113723. doi: 10.1128/mbio.01137-23. Epub 2023 Jun 21. mBio. 2023. PMID: 37341487 Free PMC article.
-
The Vi Capsular Polysaccharide of Salmonella Typhi Promotes Macrophage Phagocytosis by Binding the Human C-Type Lectin DC-SIGN.mBio. 2022 Dec 20;13(6):e0273322. doi: 10.1128/mbio.02733-22. Epub 2022 Oct 26. mBio. 2022. PMID: 36286551 Free PMC article.
-
Mechanisms of Salmonella Typhi Host Restriction.Adv Exp Med Biol. 2016;915:283-94. doi: 10.1007/978-3-319-32189-9_17. Adv Exp Med Biol. 2016. PMID: 27193549 Review.
-
Salmonella enterica serovar Typhi and the pathogenesis of typhoid fever.Annu Rev Microbiol. 2014;68:317-36. doi: 10.1146/annurev-micro-091313-103739. Annu Rev Microbiol. 2014. PMID: 25208300 Review.
Cited by
-
Virulence factors of Salmonella Typhi: interplay between the bacteria and host macrophages.Arch Microbiol. 2025 Mar 17;207(4):89. doi: 10.1007/s00203-025-04297-0. Arch Microbiol. 2025. PMID: 40095029 Review.
-
Salmonella Typhi serine threonine kinase T4519 induces lysosomal membrane permeabilization by manipulating Toll-like receptor 2-Cystatin B-Cathepsin B-NF-κB-reactive oxygen species pathway and promotes survival within human macrophages.PLoS Pathog. 2025 Apr 1;21(4):e1013041. doi: 10.1371/journal.ppat.1013041. eCollection 2025 Apr. PLoS Pathog. 2025. PMID: 40168426 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

