Cell shape sensing licenses dendritic cells for homeostatic migration to lymph nodes
- PMID: 38834865
- PMCID: PMC11224020
- DOI: 10.1038/s41590-024-01856-3
Cell shape sensing licenses dendritic cells for homeostatic migration to lymph nodes
Abstract
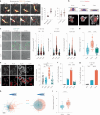
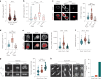
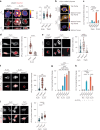
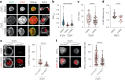
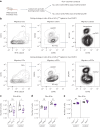
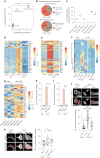
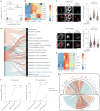
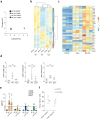
Immune cells experience large cell shape changes during environmental patrolling because of the physical constraints that they encounter while migrating through tissues. These cells can adapt to such deformation events using dedicated shape-sensing pathways. However, how shape sensing affects immune cell function is mostly unknown. Here, we identify a shape-sensing mechanism that increases the expression of the chemokine receptor CCR7 and guides dendritic cell migration from peripheral tissues to lymph nodes at steady state. This mechanism relies on the lipid metabolism enzyme cPLA2, requires nuclear envelope tensioning and is finely tuned by the ARP2/3 actin nucleation complex. We also show that this shape-sensing axis reprograms dendritic cell transcription by activating an IKKβ-NF-κB-dependent pathway known to control their tolerogenic potential. These results indicate that cell shape changes experienced by immune cells can define their migratory behavior and immunoregulatory properties and reveal a contribution of the physical properties of tissues to adaptive immunity.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
MeSH terms
Substances
Grants and funding
- SMC202006012351/Fondation pour la Recherche Médicale (Foundation for Medical Research in France)
- RGP0032-2022/Human Frontier Science Program (HFSP)
- 666003/EC | Horizon 2020 Framework Programme (EU Framework Programme for Research and Innovation H2020)
- ANR-20-CE15-0023 (InfEx)./Agence Nationale de la Recherche (French National Research Agency)
- ANR-10-IDEX-0001-02/Agence Nationale de la Recherche (French National Research Agency)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

